1EEG
 
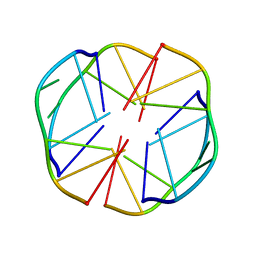 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
4HK7
 
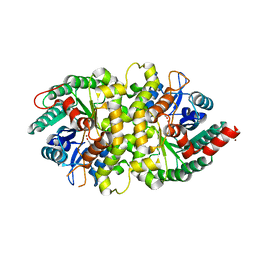 | | Crystal structure of Cordyceps militaris IDCase in complex with uracil | | Descriptor: | URACIL, Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4HK5
 
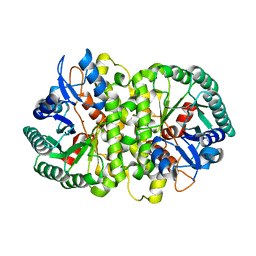 | | Crystal structure of Cordyceps militaris IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4HJW
 
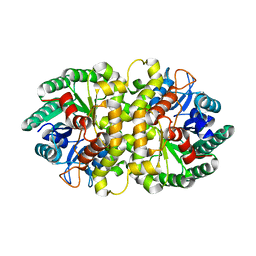 | | Crystal structure of Metarhizium anisopliae IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4IUP
 
 | |
4HK6
 
 | |
4IUU
 
 | |
4IUR
 
 | |
4IUV
 
 | |
4IUT
 
 | |
4IUQ
 
 | | crystal structure of SHH1 SAWADEE domain | | Descriptor: | CYMAL-4, SAWADEE HOMEODOMAIN HOMOLOG 1, ZINC ION | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2013-01-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1.
Nature, 498, 2013
|
|
7ES4
 
 | | the crystral structure of DndH-C-domain | | Descriptor: | DNA phosphorothioation-dependent restriction protein DptH | | Authors: | Wu, D. | | Deposit date: | 2021-05-08 | | Release date: | 2022-05-11 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The functional coupling between restriction and DNA phosphorothioate modification systems underlying the DndFGH restriction complex
Nat Catal, 2022
|
|
2HY9
 
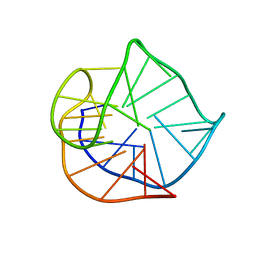 | | Human telomere DNA quadruplex structure in K+ solution hybrid-1 form | | Descriptor: | DNA (26-MER) | | Authors: | Dai, J, Punchihewa, C, Jones, R.A, Hurley, L, Yang, D. | | Deposit date: | 2006-08-04 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation.
Nucleic Acids Res., 35, 2007
|
|
6GRC
 
 | | eukaryotic junction-resolving enzyme GEN-1 binding with Sodium | | Descriptor: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
6GRB
 
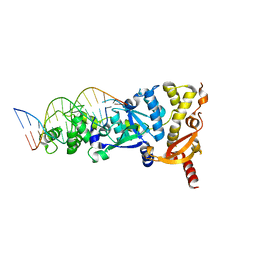 | | eukaryotic junction-resolving enzyme GEN-1 binding with Potassium | | Descriptor: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
7Z8Z
 
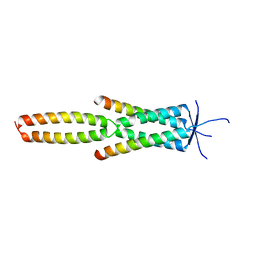 | | Crystal structure of the MEILB2-BRME1 2:2 core complex | | Descriptor: | Break repair meiotic recombinase recruitment factor 1, Heat shock factor 2-binding protein | | Authors: | Gurusaran, M, Davies, O.R. | | Deposit date: | 2022-03-19 | | Release date: | 2023-09-20 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MEILB2-BRME1 forms a V-shaped DNA clamp upon BRCA2-binding in meiotic recombination.
Nat Commun, 15, 2024
|
|
1EJZ
 
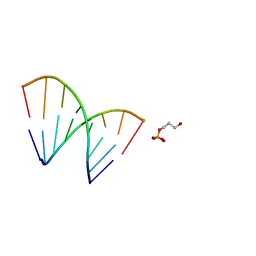 | | SOLUTION STRUCTURE OF A HNA-RNA HYBRID | | Descriptor: | DNA (5'-H(*(6HG)P*(6HC)P*(6HG)P*(6HT)P*(6HA)P*(6HG)P*(6HC)P*(6HG))-3'), PHOSPHORIC ACID MONO-(3-HYDROXY-PROPYL) ESTER, RNA (5'-R(*CP*GP*CP*UP*AP*CP*GP*C)-3') | | Authors: | Lescrnier, E, Esnouf, R, Heus, H.A, Hilbers, C.W, Herdewijn, P. | | Deposit date: | 2000-03-06 | | Release date: | 2000-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a HNA-RNA hybrid.
Chem.Biol., 7, 2000
|
|
7YT9
 
 | | crystal structure of AGD1-4 of Arabidopsis AGDP3 | | Descriptor: | AGD1-4 of Arabidopsis AGDP3 | | Authors: | Zhou, X, Du, J. | | Deposit date: | 2022-08-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The H3K9me2-binding protein AGDP3 limits DNA methylation and transcriptional gene silencing in Arabidopsis.
J Integr Plant Biol, 64, 2022
|
|
7YTA
 
 | |
8C7B
 
 | |
8AMT
 
 | | OBD-RepB pMV158 domain | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Machon, C, Amodio, J, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
8AMV
 
 | | RepB pMV158 hexamer | | Descriptor: | PHOSPHATE ION, Replication protein RepB, SODIUM ION | | Authors: | Machon, C, Amodio, J, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
7W0X
 
 | |
8C7A
 
 | |
7YF7
 
 | |
