4X8Q
 
 | | X-ray crystal structure of AlkD2 from Streptococcus mutans | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Mullins, E.A, Shi, R, Eichman, B.F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A New Family of HEAT-Like Repeat Proteins Lacking a Critical Substrate Recognition Motif Present in Related DNA Glycosylases.
Plos One, 10, 2015
|
|
6IDG
 
 | | antibody 64M-5 Fab in complex with dT(6-4)T | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), DNA (5'-D(*(64T)P*(5PY))-3') | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2018-09-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the antibody 64M-5 Fab and its complex with dT(6-4)T indicate induced-fit and high-affinity mechanisms.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6IKA
 
 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:entecavir-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6J8Y
 
 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
3V21
 
 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | DNA (5'-D(*TP*TP*CP*GP*AP*CP*CP*GP*GP*TP*CP*GP*A)-3'), Endonuclease Bse634IR | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
8SZ5
 
 | | [2T5] Self-assembling DNA motif with 5 base pairs between junctions and P32 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(P*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*CP*AP*CP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-05-26 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6HKT
 
 | | Structure of an H1-bound 6-nucleosome array | | Descriptor: | DNA (1122-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Garcia-Saez, I, Dimitrov, S, Petosa, C. | | Deposit date: | 2018-09-08 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (9.7 Å) | | Cite: | Structure of an H1-Bound 6-Nucleosome Array Reveals an Untwisted Two-Start Chromatin Fiber Conformation.
Mol. Cell, 72, 2018
|
|
6HML
 
 | | POLYADPRIBOSYL GLYCOSIDASE IN COMPLEX WITH PDD00017299 | | Descriptor: | 1-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]-6-[[(1-methylcyclopropyl)amino]-bis(oxidanyl)-$l^{4}-sulfanyl]-3-[(1-methylpyrazol-4-yl)methyl]quinazoline-2,4-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tucker, J.A, Barkauskaite, E. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
6HMM
 
 | | POLYADPRIBOSYL GLYCOHYDROLASE IN COMPLEX WITH PDD00013907 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Tucker, J.A, Brassington, C, Hassall, G. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
6HMK
 
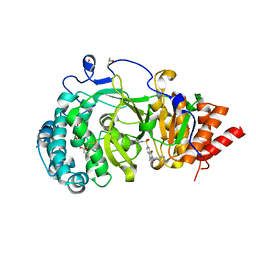 | | POLYADPRIBOSYL GLYCOHYDROLASE IN COMPLEX WITH PDD00016690 | | Descriptor: | 1-methyl-~{N}-(1-methylcyclopropyl)-3-[(2-methyl-1,3-thiazol-5-yl)methyl]-2,4-bis(oxidanylidene)quinazoline-6-sulfonamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tucker, J.A, Barkauskaite, E. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
6HMN
 
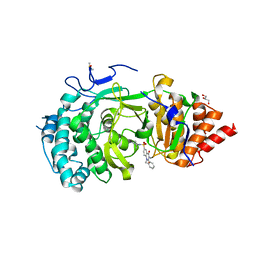 | | POLYADPRIBOSYL GLYCOSIDASE IN COMPLEX WITH PDD00014909 | | Descriptor: | 3-methyl-6-[[(1-methylcyclopropyl)amino]-bis(oxidanyl)-$l^{4}-sulfanyl]-1-(phenylmethyl)quinazoline-2,4-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tucker, J.A, Brassington, C, Hassall, G. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
7XHT
 
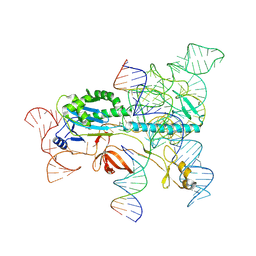 | | Structure of the OgeuIscB-omega RNA-target DNA complex | | Descriptor: | DNA (49-MER), DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3'), LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Kato, K, Okazaki, O, Isayama, Y, Ishikawa, J, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-04-10 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structure of the IscB-omega RNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9.
Nat Commun, 13, 2022
|
|
6A5K
 
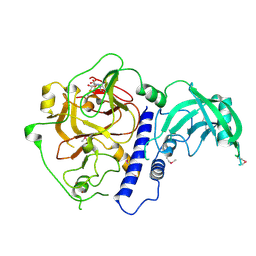 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 1 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4WHV
 
 | | E3 ubiquitin-protein ligase RNF8 in complex with Ubiquitin-conjugating enzyme E2 N and Polyubiquitin-B | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (8.3 Å) | | Cite: | RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J.Biol.Chem., 291, 2016
|
|
6JXD
 
 | |
7EUC
 
 | | Parallel G-quadruplex structure | | Descriptor: | DNA (5'-D(*TP*TP*GP*GP*GP*G)-3'), DNA (5'-D(P*GP*GP*GP*G)-3'), DNA (5'-D(P*GP*GP*GP*GP*T)-3'), ... | | Authors: | Winnerdy, F.R, Devi, G, Ang, J.C.Y, Lim, K.W, Phan, A.T. | | Deposit date: | 2021-05-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Four-Layered Intramolecular Parallel G-Quadruplex with Non-Nucleotide Loops: An Ultra-Stable Self-Folded DNA Nano-Scaffold.
Acs Nano, 16, 2022
|
|
7UYZ
 
 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
1VTF
 
 | |
3V1Z
 
 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP*G)-3'), Endonuclease Bse634IR | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
4GIG
 
 | |
3L6V
 
 | | Crystal Structure of the Xanthomonas campestris Gyrase A C-terminal Domain | | Descriptor: | DNA gyrase subunit A | | Authors: | Hsieh, T.J, Yen, T.J, Lin, T.S, Chang, H.T, Huang, S.Y, Farh, L, Chan, N.L. | | Deposit date: | 2009-12-26 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Twisting of the DNA binding surface by a beta-strand-bearing proline modulates DNA gyrase activity
To be Published
|
|
3GZ6
 
 | |
1BCE
 
 | | INTRAMOLECULAR TRIPLEX, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*AP*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*T)-3') | | Authors: | Asensio, J.L, Brown, T, Lane, A.N. | | Deposit date: | 1998-04-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of intramolecular DNA triple helices containing adjacent and non-adjacent CG.C+ triplets.
Nucleic Acids Res., 26, 1998
|
|
1JYH
 
 | |
7Y0D
 
 | |
