3TCP
 
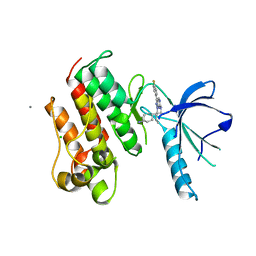 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | Descriptor: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
1IDW
 
 | | STRUCTURE OF THE HYBRID RNA/DNA R-GCUUCGGC-D[CL]U IN PRESENCE OF RH(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(UCL))-3', CHLORIDE ION, RHODIUM HEXAMINE ION | | Authors: | Cruse, W, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-04-05 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6MPT
 
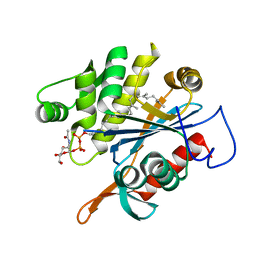 | | TagT bound to LI-WTA | | Descriptor: | 2-(acetylamino)-2-deoxy-1-O-[(S)-{[(R)-{[(2Z,6Z,10E,14E,18E)-3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-glucopyranose, CHLORIDE ION, Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagT | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2018-10-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Substrate Preferences Establish the Order of Cell Wall Assembly in Staphylococcus aureus.
J. Am. Chem. Soc., 140, 2018
|
|
1BEX
 
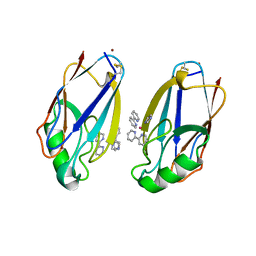 | | STRUCTURE OF RUTHENIUM-MODIFIED PSEUDOMONAS AERUGINOSA AZURIN | | Descriptor: | AZURIN, COPPER (II) ION, RUTHEMIUM BIS(2,2'-BIPYRIDINE)-2-IMIDAZOLE | | Authors: | Faham, S, Day, M.W, Rees, D.C. | | Deposit date: | 1998-05-18 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of ruthenium-modified Pseudomonas aeruginosa azurin and [Ru(2,2'-bipyridine)2(imidazole)2]SO4 x 10H2O.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1AV2
 
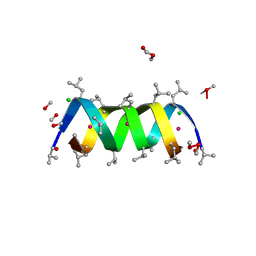 | | Gramicidin A/CsCl complex, active as a dimer | | Descriptor: | CESIUM ION, CHLORIDE ION, GRAMICIDIN A, ... | | Authors: | Burkhart, B.M, Li, N, Langs, D.A, Duax, W.L. | | Deposit date: | 1997-09-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Conducting Form of Gramicidin a is a Right-Handed Double-Stranded Double Helix.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1IHA
 
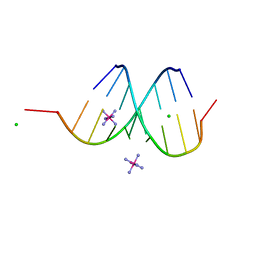 | | Structure of the Hybrid RNA/DNA R-GCUUCGGC-D[BR]U in Presence of RH(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(BRU))-3', CHLORIDE ION, RHODIUM HEXAMINE ION | | Authors: | Cruse, W.B, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4E4X
 
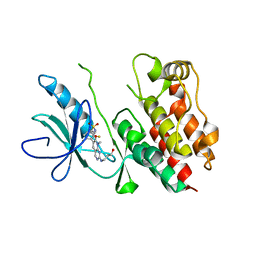 | | Crystal Structure of B-Raf Kinase Domain in Complex with a Dihydropyrido[2,3-d]pyrimidinone-based Inhibitor | | Descriptor: | N-(2,4-difluoro-3-{2-[(3-hydroxypropyl)amino]-8-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl}phenyl)propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Sturgis, H.L. | | Deposit date: | 2012-03-13 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The discovery of potent and selective pyridopyrimidin-7-one based inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1RE9
 
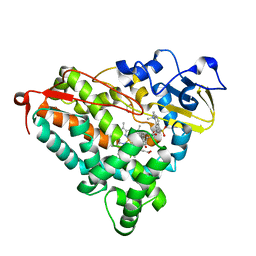 | | CRYSTAL STRUCTURE OF CYTOCHROME P450-CAM WITH A FLUORESCENT PROBE D-8-AD (ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE) | | Descriptor: | 1,2-ETHANEDIOL, ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE, Cytochrome P450-cam, ... | | Authors: | Hays, A.-M.A, Dunn, A.R, Gray, H.B, Stout, C.D, Goodin, D.B. | | Deposit date: | 2003-11-06 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational states of cytochrome P450cam revealed by trapping of synthetic molecular wires.
J.Mol.Biol., 344, 2004
|
|
1INI
 
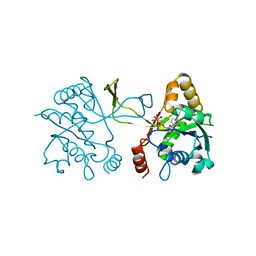 | | CRYSTAL STRUCTURE OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHETASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS, COMPLEXED WITH CDP-ME AND MG2+ | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHETASE, MAGNESIUM ION | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, A, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-05-14 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
2G5R
 
 | | Crystal structure of Siglec-7 in complex with methyl-9-(aminooxalyl-amino)-9-deoxyNeu5Ac (oxamido-Neu5Ac) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Sialic acid-binding Ig-like lectin 7, ... | | Authors: | Attrill, H, Crocker, P.R, van Aalten, D.M. | | Deposit date: | 2006-02-23 | | Release date: | 2006-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of siglec-7 in complex with sialosides: leads for rational structure-based inhibitor design.
Biochem.J., 397, 2006
|
|
2AH9
 
 | | Crystal Structure of Human M340H-Beta-1,4-Galactosyltransferase-I (M340H-B4Gal-T1) in Complex with Chitotriose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Ratner, D.M, Seeberger, P.H, Qasba, P.K. | | Deposit date: | 2005-07-27 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Preferences of beta1,4-Galactosyltransferase-I: Crystal Structures of Met340His Mutant of Human beta1,4-Galactosyltransferase-I with a Pentasaccharide and Trisaccharides of the N-Glycan Moiety
J.Mol.Biol., 353, 2005
|
|
1UJW
 
 | | Structure of the complex between BtuB and Colicin E3 Receptor binding domain | | Descriptor: | 2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 3-OXO-PENTADECANOIC ACID, ACETOACETIC ACID, ... | | Authors: | Kurisu, G, Zakharov, S.D, Zhalnina, M.V, Bano, S, Eroukova, V.Y, Rokitskaya, T.I, Antonenko, Y.N, Wiener, M.C, Cramer, W.A. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of BtuB with bound colicin E3 R-domain implies a translocon
NAT.STRUCT.BIOL., 10, 2003
|
|
3ECB
 
 | | Crystal structure of mouse H-2Dd in complex with peptide P18-I10 derived from human immunodeficiency virus envelope glycoprotein 120 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2 microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Natarajan, K, Wang, R, Margulies, D.H. | | Deposit date: | 2008-08-29 | | Release date: | 2009-07-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structural basis of the CD8alphabeta/MHC class i interaction: focused recognition orients CD8beta to a T cell proximal position
J.Immunol., 183, 2009
|
|
2QV4
 
 | | Human pancreatic alpha-amylase complexed with nitrite and acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5R,6S)-4-{[alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl]oxy}-5,6-dihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-08-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
3EJ7
 
 | | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity | | Descriptor: | Alpha-subunit of trans-3-chloroacrylic acid dehalogenase, Beta-subunit of trans-3-chloroacrylic acid dehalogenase, SULFATE ION | | Authors: | Pegan, S, Serrano, H, Whitman, C.P, Mesecar, A.D. | | Deposit date: | 2008-09-17 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3QPE
 
 | | Crystal structure of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with D-Galacturonate and 5-keto-4-deoxy-D-Galacturonate | | Descriptor: | 4-deoxy-L-threo-hex-5-ulosuronic acid, D-galacturonic acid, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Mills-Groninger, F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-12 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal structure of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with D-Galacturonate and 5-keto-4-deoxy-D-Galacturonate
To be Published
|
|
1UL3
 
 | | Crystal Structure of PII from Synechocystis sp. PCC 6803 | | Descriptor: | CALCIUM ION, GLYCEROL, Nitrogen regulatory protein P-II | | Authors: | Xu, Y, Carr, P.D, Clancy, P, Garcia-Dominguez, M, Forchhammer, K, Florencio, F, Tandeau de Marsac, N, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 2003-09-09 | | Release date: | 2003-12-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the PII proteins from the cyanobacteria Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1DR9
 
 | | CRYSTAL STRUCTURE OF A SOLUBLE FORM OF B7-1 (CD80) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T LYMPHOCYTE ACTIVATION ANTIGEN | | Authors: | Ikemizu, S, Jones, E.Y, Stuart, D.I, Davis, S.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-10 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and dimerization of a soluble form of B7-1.
Immunity, 12, 2000
|
|
4CC0
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN JAGGED-1, ... | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4KOA
 
 | | Crystal Structure Analysis of 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti | | Descriptor: | 1,5-anhydro-D-fructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schu, M, Faust, A, Stosik, B, Kohring, G.-W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2013-05-11 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structure of substrate-free 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti 1021 reveals an open enzyme conformation.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4KOL
 
 | | The structure of hemagglutinin from avian-origin H7N9 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2 | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses.
Science, 342, 2013
|
|
4KFQ
 
 | | Crystal structure of the NMDA receptor GluN1 ligand binding domain in complex with 1-thioxo-1,2-dihydro-[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one | | Descriptor: | 1-sulfanyl[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Steffensen, T.B, Tabrizi, F.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-04-27 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site.
J.Biol.Chem., 288, 2013
|
|
1MVE
 
 | | Crystal structure of a natural circularly-permutated jellyroll protein: 1,3-1,4-beta-D-glucanase from Fibrobacter succinogenes | | Descriptor: | CALCIUM ION, Truncated 1,3-1,4-beta-D-glucanase | | Authors: | Tsai, L.-C, Shyur, L.-F, Lee, S.-H, Lin, S.-S, Yuan, H.S. | | Deposit date: | 2002-09-25 | | Release date: | 2003-07-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Natural Circularly Permuted Jellyroll Protein: 1,3-1,4-beta-D-Glucanase from Fibrobacter succinogenes.
J.Mol.Biol., 330, 2003
|
|
2QMK
 
 | | Human pancreatic alpha-amylase complexed with nitrite | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRITE ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-07-16 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
309D
 
 | |
