6C75
 
 | |
6C7L
 
 | | Structure of Iron containing alcohol dehydrogenase from Thermococcus thioreducens in a tetragonal crystal form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jones, J.A, Larson, S.B, McPherson, A. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structure of an iron-containing alcohol dehydrogenase from a hyperthermophilic archaeon in two chemical states.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
1P59
 
 | | Structure of a non-covalent Endonuclease III-DNA Complex | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*GP*(5IU)P*GP*GP*AP*C)-3', 5'-D(TP*GP*(5IU)P*CP*CP*AP*(3DR)P*GP*(5IU)P*CP*T)-3', Endonuclease III, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2003-04-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Trapped Endonuclease III-DNA Covalent Intermediate
Embo J., 22, 2003
|
|
6HO9
 
 | | TRANSCRIPTIONAL REPRESSOR ETHR FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH BDM44825 | | Descriptor: | 4-[4-(2-methylpropyl)-1,2,3-triazol-1-yl]-~{N}-[3,3,3-tris(fluoranyl)propyl]benzamide, HTH-type transcriptional regulator EthR | | Authors: | Wintjens, R, Wohlkonig, A. | | Deposit date: | 2018-09-17 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A comprehensive analysis of the protein-ligand interactions in crystal structures of Mycobacterium tuberculosis EthR.
Biochim Biophys Acta Proteins Proteom, 1867, 2018
|
|
2Q5R
 
 | | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase | | Descriptor: | Tagatose-6-phosphate kinase | | Authors: | McGrath, T.E, Soloveychik, M, Romanov, V, Thambipillai, D, Dharamsi, A, Virag, C, Domagala, M, Pai, E.F, Edwards, A.M, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase
TO BE PUBLISHED
|
|
6C76
 
 | |
2HMN
 
 | | Crystal Structure of the Naphthalene 1,2-Dioxygenase F352V Mutant Bound to Anthracene. | | Descriptor: | 1,2-ETHANEDIOL, ANTHRACENE, FE (III) ION, ... | | Authors: | Ferraro, D.J, Okerlund, A.L, Mowers, J.C, Ramaswamy, S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
J.Bacteriol., 188, 2006
|
|
3BT6
 
 | | Crystal Structure of Influenza B Virus Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q, Cheng, F, Lu, M, Tian, X, Ma, J. | | Deposit date: | 2007-12-27 | | Release date: | 2008-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of unliganded influenza B virus hemagglutinin.
J.Virol., 82, 2008
|
|
5WTR
 
 | | Crystal structure of a prokaryotic TRIC channel in 0.5 M KCl | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, TRISTEAROYLGLYCEROL, ... | | Authors: | Ou, X.M, Wang, L.F, Yang, H.T, Liu, X.Y, Liu, Z.F. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ion and water binding sites inside an occluded hourglass pore of a TRIC channel
BMC BIOL., 2017
|
|
7GSL
 
 | |
7GSM
 
 | |
7GT0
 
 | |
7GTA
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000065a | | Descriptor: | (5S)-N-(4-fluorophenyl)-5-methyl-4,5-dihydro-1,3-thiazol-2-amine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTH
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000637a | | Descriptor: | (6aR,8R,12R,12aS)-2-methyl-6a,10,11,12a-tetrahydro-6H,7H,9H-[1]benzopyrano[4,3-c]pyrazolo[1,2-a]pyrazol-9-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
3NFS
 
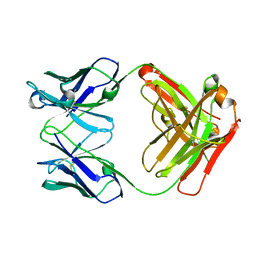 | | Crystal structure the Fab fragment of therapeutic antibody daclizumab | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
2ZH6
 
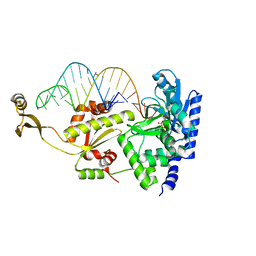 | | Complex structure of AFCCA with tRNAminiDCU and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CCA-adding enzyme, MAGNESIUM ION, ... | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2008-02-01 | | Release date: | 2008-08-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme
Embo J., 27, 2008
|
|
2ZH4
 
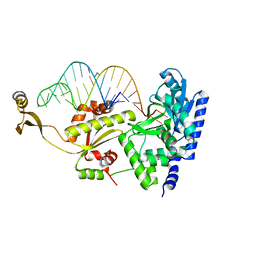 | | Complex structure of AFCCA with tRNAminiDCG | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (34-MER) | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2008-02-01 | | Release date: | 2008-08-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme
Embo J., 27, 2008
|
|
6L02
 
 | |
2KA5
 
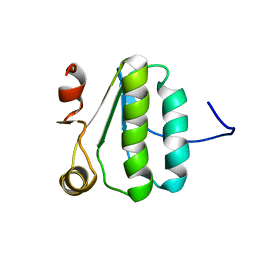 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4ZAU
 
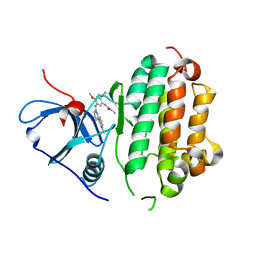 | | AZD9291 complex with wild type EGFR | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Squire, C.J, Yosaatmadja, Y, Flanagan, J.U, McKeage, M. | | Deposit date: | 2015-04-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed.
J.Struct.Biol., 192, 2015
|
|
7GTL
 
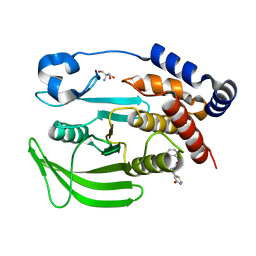 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000554a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, benzyl (3aS,8aS)-1-oxooctahydropyrrolo[3,4-d]azepine-6(1H)-carboxylate | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSD
 
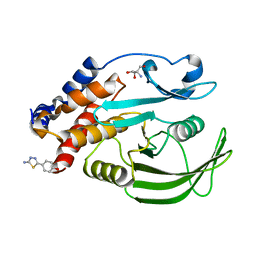 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000605a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(5-amino-1,3,4-thiadiazol-2-yl)phenol, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSO
 
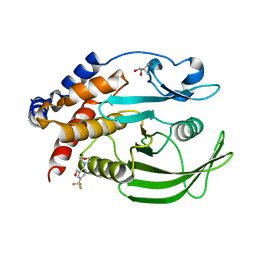 | |
7GSW
 
 | |
7GSF
 
 | |
