3R0G
 
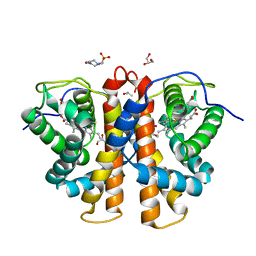 | | 3D Structure of Ferric Methanosarcina Acetivorans Protoglobin I149F mutant in Aquomet form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
8U4L
 
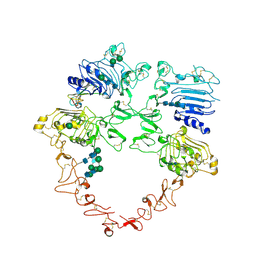 | | Structure of the HER2/HER4/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
5NMC
 
 | |
6IM1
 
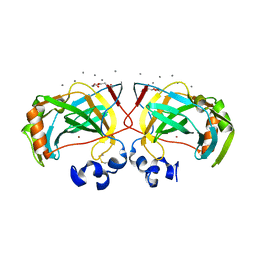 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), TETRAETHYLENE GLYCOL, ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
8U4K
 
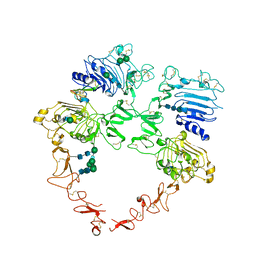 | | Structure of the HER2/HER4/BTC Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U78
 
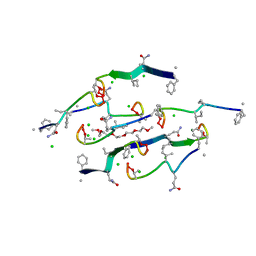 | | Structure of a N-Me-D-Gln4,Lys10-teixobactin analogue | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, N-Methyl-D-Gln4,Lys10-teixobactin | | Authors: | Nowick, J.S, Yang, H, Kreutzer, A.G. | | Deposit date: | 2023-09-14 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Interactions of Teixobactin Analogues in the Crystal State.
J.Org.Chem., 89, 2024
|
|
6H0F
 
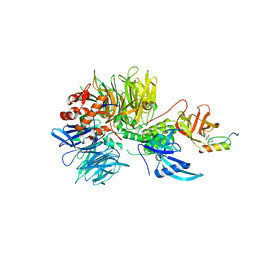 | | Structure of DDB1-CRBN-pomalidomide complex bound to IKZF1(ZF2) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, DNA-binding protein Ikaros, Protein cereblon, ... | | Authors: | Petzold, G, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
5J85
 
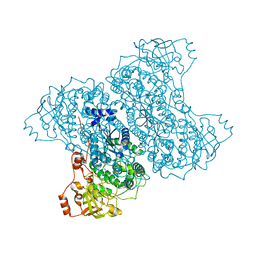 | | Ser480Ala mutant of L-arabinonate dehydratase | | Descriptor: | Dihydroxyacid dehydratase/phosphogluconate dehydratase, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION | | Authors: | Rahman, M.M, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2016-04-07 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of a Bacterial l-Arabinonate Dehydratase Contains a [2Fe-2S] Cluster.
ACS Chem. Biol., 12, 2017
|
|
5N76
 
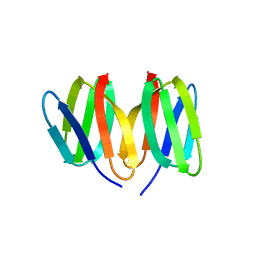 | | Crystal structure of the apo-form of the CO dehydrogenase accessory protein CooT from Rhodospirillum rubrum | | Descriptor: | CooT | | Authors: | Timm, J, Brochier-Armanet, C, Perard, J, Zambelli, B, Ollagnier-de-Choudens, S, Ciurli, S, Cavazza, C. | | Deposit date: | 2017-02-19 | | Release date: | 2017-05-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The CO dehydrogenase accessory protein CooT is a novel nickel-binding protein.
Metallomics, 9, 2017
|
|
4WQ5
 
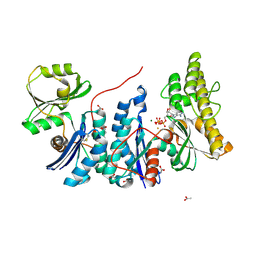 | | YgjD(V85E)-YeaZ heterodimer in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, FE (III) ION, ... | | Authors: | Zhang, W. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
7XYH
 
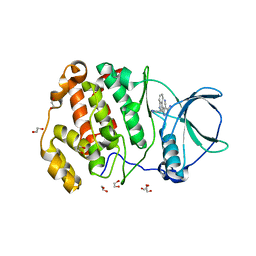 | | Crystal structure of CK2a2 complexed with AG1112 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-3-[(~{Z})-1-cyano-2-(1~{H}-indol-3-yl)ethenyl]-1~{H}-pyrazole-4-carbonitrile, Casein kinase II subunit alpha' | | Authors: | Ikeda, A, Kinoshita, T, Tsuyuguchi, M. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Bivalent binding mode of an amino-pyrazole inhibitor indicates the potentials for CK2 alpha 1-selective inhibitors.
Biochem.Biophys.Res.Commun., 630, 2022
|
|
5NG0
 
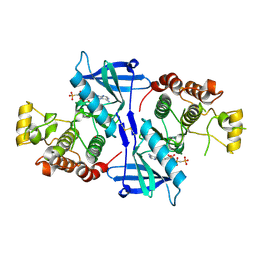 | | Structure of RIP2K(L294F) with bound AMPPCP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
5NG3
 
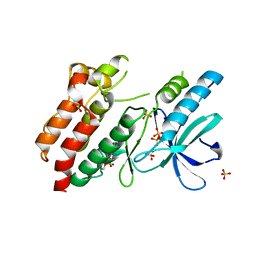 | | Structure of inactive kinase RIP2K(K47R) | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, SULFATE ION | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
6OWV
 
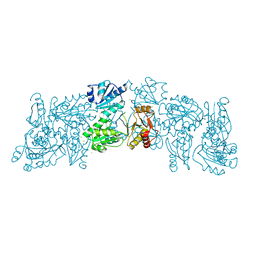 | | Crystal structure of a Human Cardiac Calsequestrin Filament | | Descriptor: | CHLORIDE ION, Calsequestrin-2, SULFATE ION | | Authors: | Titus, E.W, Deiter, F.H, Shi, C, Jura, N, Deo, R.C. | | Deposit date: | 2019-05-12 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The structure of a calsequestrin filament reveals mechanisms of familial arrhythmia.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7B69
 
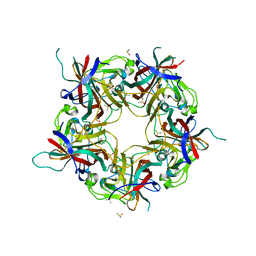 | |
7B6A
 
 | | BK Polyomavirus VP1 pentamer core (residues 30-299) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Osipov, E.M, Munawar, A, Beelen, S, Strelkov, S.V. | | Deposit date: | 2020-12-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
3KI8
 
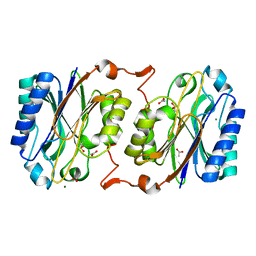 | | Crystal structure of hyperthermophilic nitrilase | | Descriptor: | ACETIC ACID, Beta ureidopropionase (Beta-alanine synthase), MAGNESIUM ION | | Authors: | Raczynska, J, Vorgias, C, Antranikian, G, Rypniewski, W. | | Deposit date: | 2009-10-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic analysis of a thermoactive nitrilase.
J.Struct.Biol., 173, 2010
|
|
4WQM
 
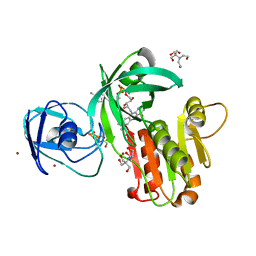 | | Structure of the toluene 4-monooxygenase NADH oxidoreductase T4moF, K270S K271S variant | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Acheson, J.F, Fox, B.G. | | Deposit date: | 2014-10-22 | | Release date: | 2015-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of T4moF, the Toluene 4-Monooxygenase Ferredoxin Oxidoreductase.
Biochemistry, 54, 2015
|
|
7B6C
 
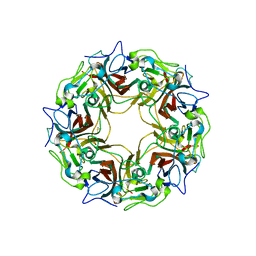 | | BK Polyomavirus VP1 pentamer fusion with long C-terminal extended arm | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Major capsid protein VP1,Major capsid protein VP1 | | Authors: | Osipov, E.M, Beelen, S, Strelkov, S.V. | | Deposit date: | 2020-12-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
6OWW
 
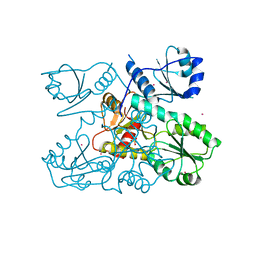 | | Crystal structure of a Human Cardiac Calsequestrin Filament Complexed with Ytterbium | | Descriptor: | Calsequestrin-2, SULFATE ION, YTTERBIUM (III) ION | | Authors: | Titus, E.W, Deiter, F.H, Shi, C, Jura, N, Deo, R.C. | | Deposit date: | 2019-05-12 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | The structure of a calsequestrin filament reveals mechanisms of familial arrhythmia.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3S8G
 
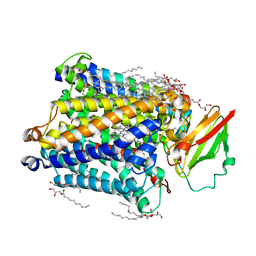 | | 1.8 A structure of ba3 cytochrome c oxidase mutant (A120F) from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
6OY3
 
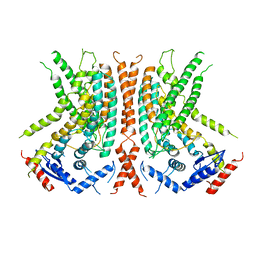 | | nhTMEM16 L302A +Ca2+ in nanodiscs | | Descriptor: | CALCIUM ION, nhTMEM16 | | Authors: | Falzone, M, Lee, B.C, Accardi, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Dynamic modulation of the lipid translocation groove generates a conductive ion channel in Ca 2+ -bound nhTMEM16.
Nat Commun, 10, 2019
|
|
6H0G
 
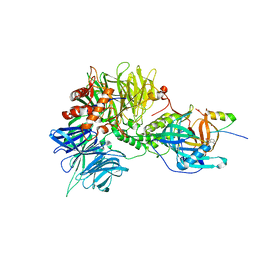 | | Structure of the DDB1-CRBN-pomalidomide complex bound to ZNF692(ZF4) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DDB1 (DNA damage binding protein 1),DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Bunker, R.D, Petzold, G, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
3KLC
 
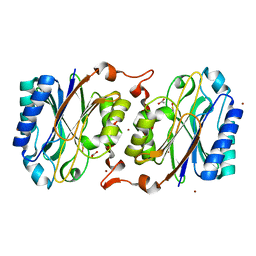 | | Crystal structure of hyperthermophilic nitrilase | | Descriptor: | ACETIC ACID, BROMIDE ION, Beta ureidopropionase (Beta-alanine synthase), ... | | Authors: | Raczynska, J, Vorgias, C, Antranikian, G, Rypniewski, W. | | Deposit date: | 2009-11-07 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystallographic analysis of a thermoactive nitrilase.
J.Struct.Biol., 173, 2010
|
|
6P7A
 
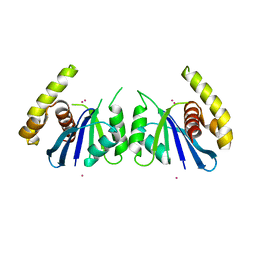 | | CRYSTAL STRUCTURE OF THE FOWLPOX VIRUS HOLLIDAY JUNCTION RESOLVASE | | Descriptor: | CADMIUM ION, Holliday junction resolvase | | Authors: | Li, N, Shi, K, Banerjee, S, Rao, T, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.081 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
