8JNO
 
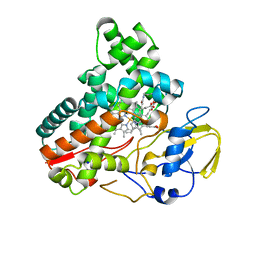 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-deOH-HSAF | | Descriptor: | (1Z,3E,5S,7S,8R,9S,10S,11R,13R,15R,16S,18Z,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3T7Z
 
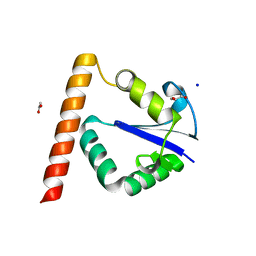 | | Structure of Methanocaldococcus jannaschii Nop N-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleolar protein Nop 56/58, ... | | Authors: | Biswas, S, Maxwell, S. | | Deposit date: | 2011-07-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structurally Conserved Nop56/58 N-terminal Domain Facilitates Archaeal Box C/D Ribonucleoprotein-guided Methyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
3SPX
 
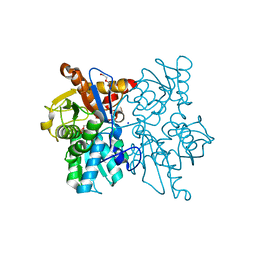 | | Crystal structure of O-Acetyl Serine Sulfhydrylase from Leishmania donovani | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, O-acetyl serine sulfhydrylase, ... | | Authors: | Raj, I, Gourinath, S. | | Deposit date: | 2011-07-04 | | Release date: | 2012-07-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The narrow active-site cleft of O-acetylserine sulfhydrylase from Leishmania donovani allows complex formation with serine acetyltransferases with a range of C-terminal sequences
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SVT
 
 | |
8J2L
 
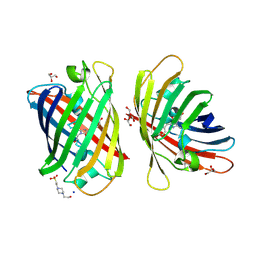 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutations (N137A, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2H
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (N137A) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | GLYCEROL, SODIUM ION, StayGold(N137A) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
1XSS
 
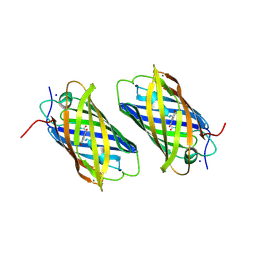 | | Semi-rational engineering of a green-emitting coral fluorescent protein into an efficient highlighter. | | Descriptor: | MAGNESIUM ION, SODIUM ION, fluorescent protein | | Authors: | Tsutsui, H, Karasawa, S, Shimizu, H, Nukina, N, Miyawaki, A. | | Deposit date: | 2004-10-20 | | Release date: | 2005-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Semi-rational engineering of a coral fluorescent protein into an efficient highlighter
Embo Rep., 6, 2005
|
|
3T3F
 
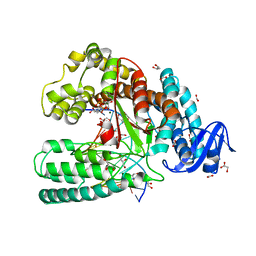 | | Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and dNITP | | Descriptor: | 1-{2-DEOXY-5-O-[(R)-HYDROXY{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}PHOSPHORYL]-BETA-D-ERYTHRO-PENTOFURANOSYL}-5-NITRO -1H-INDOLE, 5'-D(*AP*AP*AP*(3DR)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3', ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amino acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase.
J.Biol.Chem., 287, 2012
|
|
1XSN
 
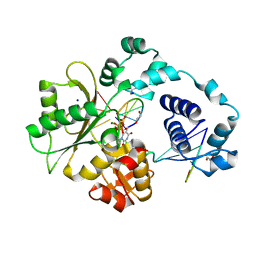 | | Crystal Structure of human DNA polymerase lambda in complex with a one nucleotide DNA gap and ddTTP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*(2DT))-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
8JP0
 
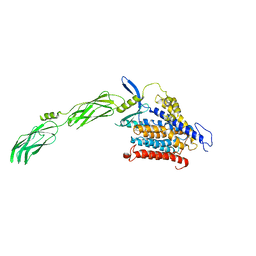 | | structure of human sodium-calciumexchanger NCX1 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, Sodium/calcium exchanger 1 | | Authors: | Dong, Y, Zhao, Y. | | Deposit date: | 2023-06-09 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the allosteric inhibition of human sodium-calcium exchanger NCX1 by XIP and SEA0400.
Embo J., 43, 2024
|
|
3PKT
 
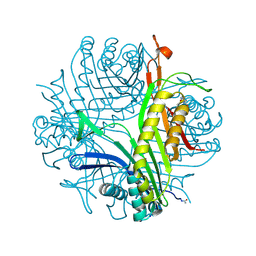 | | Urate oxidase under 2 MPa / 20 bars pressure of nitrous oxide | | Descriptor: | 8-AZAXANTHINE, NITROUS OXIDE, SODIUM ION, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-12 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
3PO4
 
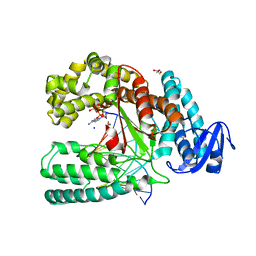 | | Structure of a mutant of the large fragment of DNA polymerase I from Thermus aquaticus in complex with a blunt-ended DNA and ddATP | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3'), DNA (5'-D(*TP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Learning from Directed Evolution: Thermus aquaticus DNA Polymerase Mutants with Translesion Synthesis Activity.
Chembiochem, 12, 2011
|
|
3Q3U
 
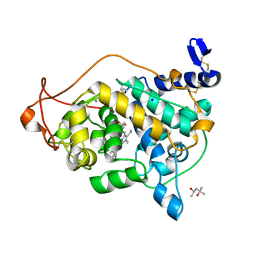 | | Trametes cervina lignin peroxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Calvino, F.R, Romero, A, Miki, Y, Martinez, A.T. | | Deposit date: | 2010-12-22 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic, kinetic, and spectroscopic study of the first ligninolytic peroxidase presenting a catalytic tyrosine.
J.Biol.Chem., 286, 2011
|
|
3PKS
 
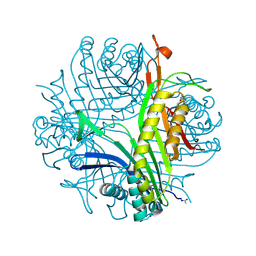 | | Urate oxidase under 1.5 MPa / 15 bars pressure of nitrous oxide | | Descriptor: | 8-AZAXANTHINE, NITROUS OXIDE, SODIUM ION, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-12 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
3PLI
 
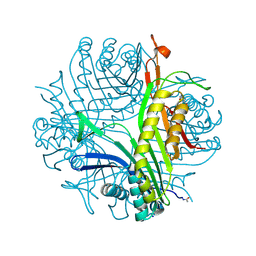 | | Urate oxidase under 1.8 MPa / 18 bars pressure of equimolar mixture xenon : nitrous oxide | | Descriptor: | 8-AZAXANTHINE, NITROUS OXIDE, SODIUM ION, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-15 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
1YPE
 
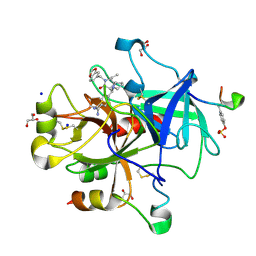 | | Thrombin Inhibitor Complex | | Descriptor: | (1R,3AS,4R,8AS,8BR)-4-(2-BENZO[1,3]DIOXOL-5-YLMETHYL-1-ETHYL-3-OXO-DECAHYDRO-PYRROLO[3,4-A]PYRROLIZIN-4-YL)-BENZAMIDINE, GLYCEROL, Hirudin, ... | | Authors: | Fokkens, J, Obst-Sander, U, Heine, A, Diederich, F, Klebe, G. | | Deposit date: | 2005-01-31 | | Release date: | 2006-01-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A simple protocol to estimate differences in protein binding affinity for enantiomers without prior resolution of racemates
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
3PK3
 
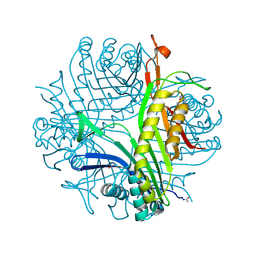 | | urate oxidase under 3.0 MPa / 30 bars pressure of nitrous oxide | | Descriptor: | 8-AZAXANTHINE, NITROUS OXIDE, SODIUM ION, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
3PKF
 
 | | Urate oxidase under 0.2 MPa / 2 bars pressure of equimolar mixture of xenon and nitrous oxide | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
3PLH
 
 | | urate oxidase under 1.5 MPa / 15 bars pressure of equimolar mixture xenon : nitrous oxide | | Descriptor: | 8-AZAXANTHINE, NITROUS OXIDE, SODIUM ION, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-15 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
8IZJ
 
 | |
1ZPK
 
 | | Crystal structure of the complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[R-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | CHLORIDE ION, N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN, ... | | Authors: | Duskova, J, Skalova, T, Dohnalek, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-17 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mutational Study of Pseudopeptide Inhibitor Binding to HIV-1 Protease; Analysis of Four X-ray Structures
To be Published
|
|
3TH4
 
 | | Mg2+ Is Required for Optimal Folding of the Gamma-Carboxyglutamic Acid (Gla) Domains of Vitamin K-Dependent Clotting Factors At Physiological Ca2+ | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII heavy chain, ... | | Authors: | Vadivel, K, Agah, S, Cascio, D, Padmanabhan, K, Bajaj, S.P. | | Deposit date: | 2011-08-18 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mg2+ Is Required for Optimal Folding of the Gamma-Carboxyglutamic Acid (Gla)
Domains of Vitamin K-Dependent Clotting Factors At Physiological Ca2+
To be Published
|
|
8J0I
 
 | | Aldo-keto reductase KmAKR | | Descriptor: | NADPH-dependent alpha-keto amide reductase, SODIUM ION | | Authors: | Xu, S.Y, Zhou, L, Xu, Y, Wang, Y.J, Zheng, Y.G. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Aldo-keto reductase KmAKR from Kluyveromyces marxianus
To Be Published
|
|
1JG3
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | Descriptor: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1ZQT
 
 | |
