5MCN
 
 | | Radiation damage to GH7 Family Cellobiohydrolase from Daphnia pulex: Dose (DWD) 22.7 MGy | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL, SULFATE ION | | Authors: | Bury, C.S, McGeehan, J.E, Ebrahim, A, Garman, E.F. | | Deposit date: | 2016-11-10 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | OH cleavage from tyrosine: debunking a myth.
J Synchrotron Radiat, 24, 2017
|
|
5ME5
 
 | | Crystal Structure of eiF4E from C. melo bound to a eIF4G peptide | | Descriptor: | Eukaryotic transcription initiation factor 4E, SULFATE ION, eIF4G | | Authors: | Querol-Audi, J, Silva, C, Miras, M, Truniger, V, Aranda-Regules, M, Verdaguer, N. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of eIF4E in Complex with an eIF4G Peptide Supports a Universal Bipartite Binding Mode for Protein Translation.
Plant Physiol., 174, 2017
|
|
4QBZ
 
 | | Crystal structure of human TLR8 in complex with DS-802 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-butyl[1,3]oxazolo[4,5-c]quinolin-4-amine, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of Activity at Human Toll-like Receptors 7 and 8: Quantitative Structure-Activity Relationship (QSAR) of Diverse Heterocyclic Scaffolds
J.Med.Chem., 57, 2014
|
|
9G0F
 
 | | CryoEM structure of PmcTnsC-dsDNA-AMPPNP | | Descriptor: | AAA+ ATPase domain-containing protein, DNA, MAGNESIUM ION, ... | | Authors: | Finocchio, G, Chanez, C, Querques, I, Speichert, K.J, Jinek, M. | | Deposit date: | 2024-07-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of TnsC oligomerization and transposase recruitment in type I-B CRISPR-associated transposons.
Nucleic Acids Res., 53, 2025
|
|
6H5T
 
 | | Intersectin SH3A short isoform | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Driller, J.H, Gerth, F, Freund, C, Wahl, M.C, Loll, B. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Exon Inclusion Modulates Conformational Plasticity and Autoinhibition of the Intersectin 1 SH3A Domain.
Structure, 27, 2019
|
|
6OWR
 
 | | NMR solution structure of YfiD | | Descriptor: | Autonomous glycyl radical cofactor | | Authors: | Bowman, S.E.J, Drennan, C.L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme.
J.Biol.Inorg.Chem., 24, 2019
|
|
8QUD
 
 | | Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5 | | Descriptor: | (5R)-5-ethyl-3-(6-spiro[2H-1-benzofuran-3,1'-cyclopropane]-4-yloxypyridin-3-yl)imidazolidine-2,4-dione, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, G, Mckinley, G, Marsden, B, Pike, A.C.W, Ye, M, Brooke, L.M, Bakshi, S, Lakshminarayana, B, Pilati, N, Marasco, A, Gunthorpe, M, Alvaro, G.S, Large, C.H, Williams, E, Sauer, D.B. | | Deposit date: | 2023-10-16 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The binding and mechanism of a positive allosteric modulator of Kv3 channels.
Nat Commun, 15, 2024
|
|
3HW7
 
 | | High pressure (0.57 GPa) crystal structure of bovine copper, zinc superoxide dismutase at 2.0 angstroms | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Ascone, I, Savino, C. | | Deposit date: | 2009-06-17 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of the Cu,Zn superoxide dismutase structure investigated at 0.57 GPa
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6WMB
 
 | |
7PJS
 
 | | Structure of the 70S ribosome with tRNAs in the classical pre-translocation state and apramycin (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement
Nat Commun, 12, 2021
|
|
5WOP
 
 | | High Resolution Structure of Mutant CA09-PB2cap | | Descriptor: | GLYCEROL, Polymerase PB2 | | Authors: | Constantinides, A.E, Gumpper, R.H, Severin, C, Luo, M. | | Deposit date: | 2017-08-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | High-resolution structure of the Influenza A virus PB2cap binding domain illuminates the changes induced by ligand binding.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7ZHS
 
 | | 3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F by imposing D5 symmetry | | Descriptor: | Ubiquitin-like protein SMT3,DnaJ homolog subfamily A member 2, ZINC ION | | Authors: | Cuellar, J, Velasco-Carneros, L, Santiago, C, Martin-Benito, J, Valpuesta, J, Muga, A. | | Deposit date: | 2022-04-07 | | Release date: | 2023-07-26 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The self-association equilibrium of DNAJA2 regulates its interaction with unfolded substrate proteins and with Hsc70.
Nat Commun, 14, 2023
|
|
1VYX
 
 | | Solution structure of the KSHV K3 N-terminal domain | | Descriptor: | ORF K3, ZINC ION | | Authors: | Dodd, R.B, Allen, M.D, Brown, S.E, Sanderson, C.M, Duncan, L.M, lehner, P.J, Bycroft, M, Read, R.J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-10-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Kaposi'S Sarcoma-Associated Herpesvirus K3 N-Terminal Domain Reveals a Novel E2-Binding C4Hc3-Type Ring Domain
J.Biol.Chem., 279, 2004
|
|
2XD8
 
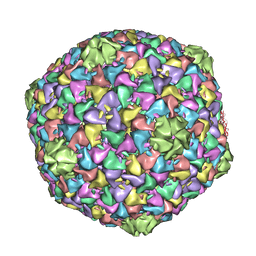 | | Capsid structure of the infectious Prochlorococcus Cyanophage P-SSP7 | | Descriptor: | T7-LIKE CAPSID PROTEIN | | Authors: | Liu, X, Zhang, Q, Murata, K, Baker, M.L, Sullivan, M.B, Fu, C, Dougherty, M, Schmid, M.F, Osburne, M.S, Chisholm, S.W, Chiu, W. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Changes in a Marine Podovirus Associated with Release of its Genome Into Prochlorococcus
Nat.Struct.Mol.Biol., 17, 2010
|
|
9DMM
 
 | | Crystal structure of human KRAS G12C covalently bound to Divarasib (GDC6036) | | Descriptor: | 1-{(3S)-4-[(7M)-7-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}quinazolin-4-yl]-3-methylpiperazin-1-yl}propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Fernando, M.C. | | Deposit date: | 2024-09-13 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of KRAS G12C bound to divarasib highlights features of potent switch-II pocket engagement.
Small Gtpases, 15, 2024
|
|
7ZG3
 
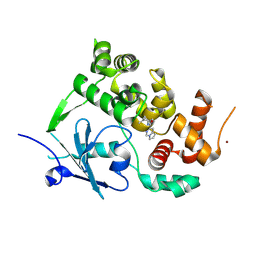 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228 | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-[(1~{S})-1,2,2-trimethylcyclopropyl]pyrrolo[1,2-c]pyrimidine-3-carboxamide | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-04-01 | | Release date: | 2023-08-16 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
4RE0
 
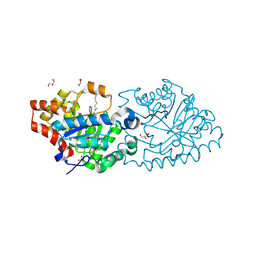 | | Crystal structure of VmoLac in P622 space group | | Descriptor: | COBALT (II) ION, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Hiblot, J, Bzdrenga, J, Champion, C, Gotthard, G, Gonzalez, D, Chabriere, E, Elias, M. | | Deposit date: | 2014-09-20 | | Release date: | 2015-02-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia.
Sci Rep, 5, 2015
|
|
6R83
 
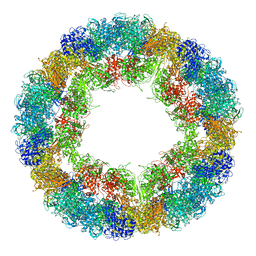 | | CryoEM structure and molecular model of squid hemocyanin (Todarodes pacificus , TpH) | | Descriptor: | Hemocyanin subunit 1 | | Authors: | Tanaka, Y, Kato, S, Stabrin, M, Raunser, S, Matsui, T, Gatsogiannis, C. | | Deposit date: | 2019-03-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryo-EM reveals the asymmetric assembly of squid hemocyanin.
Iucrj, 6, 2019
|
|
4WWF
 
 | | High-resolution structure of two Ni-bound forms of the M123C mutant of C. metallidurans CnrXs | | Descriptor: | NICKEL (II) ION, Nickel and cobalt resistance protein CnrR, SODIUM ION | | Authors: | Volbeda, A, Coves, J, Maillard, A.P, Kinnemann, S, Grosse, C, Schleuder, G, Petit-Hurtlein, I, de Rosny, E, Nies, D.H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
8Z0P
 
 | | Cryo-EM structure of human ELAC2 | | Descriptor: | PHOSPHATE ION, ZINC ION, Zinc phosphodiesterase ELAC protein 2 | | Authors: | Liu, Z.M, Xue, C.Y. | | Deposit date: | 2024-04-10 | | Release date: | 2024-12-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human ELAC2 as a tRNA 3' processing enzyme.
Nucleic Acids Res., 52, 2024
|
|
2XDF
 
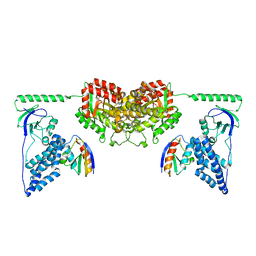 | | Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | Authors: | Schwieters, C.D, Suh, J.-Y, Grishaev, A, Guirlando, R, Takayama, Y, Clore, G.M. | | Deposit date: | 2010-04-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of the 128 kDa Enzyme I Dimer from Escherichia Coli and its 146 kDa Complex with Hpr Using Residual Dipolar Couplings and Small- and Wide-Angle X-Ray Scattering.
J.Am.Chem.Soc., 132, 2010
|
|
8CX0
 
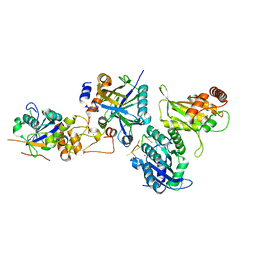 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC monomeric complex | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
6Q6G
 
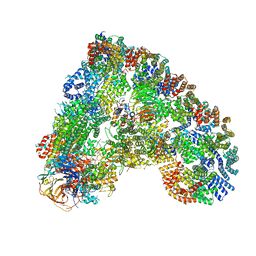 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Zhang, S, Barford, D. | | Deposit date: | 2018-12-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
6UCX
 
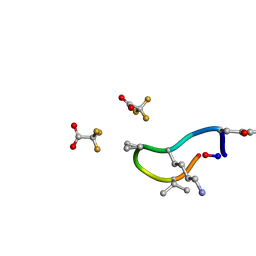 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6GO6
 
 | | TdT chimera (Loop1 of pol mu) - ternary complex with downstream dsDNA | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*AP*AP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*GP*GP*C)-3'), ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
