6T5X
 
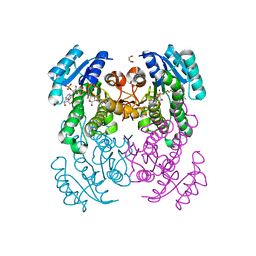 | | Crystal structure of Salmonella typhimurium FabG in complex with NADPH at 1.5 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vella, P, Schnell, R, Schneider, G. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
6TOM
 
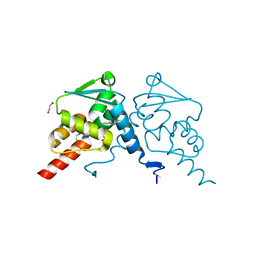 | | Crystal structure of human BCL6 BTB domain in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[2-[(3~{S},5~{R})-4,4-bis(fluoranyl)-3,5-dimethyl-piperidin-1-yl]-5-chloranyl-pyrimidin-4-yl]amino]-1-methyl-3-(3-methyl-3-oxidanyl-butyl)benzimidazol-2-one, B-cell lymphoma 6 protein | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
6T62
 
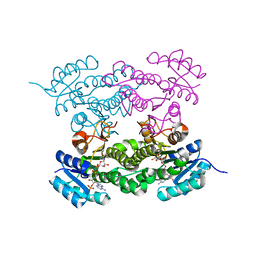 | |
7QLD
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I, Co-crystallization with HgCl2, Mutation Ser146Cys, (aa 32-198) | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, S-layer protein | | Authors: | Sagmeister, T, Vejzovic, D, Eder, M, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLE
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I (aa 32-198) | | Descriptor: | S-layer protein | | Authors: | Sagmeister, T, Eder, M, Vejzovic, D, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3RQP
 
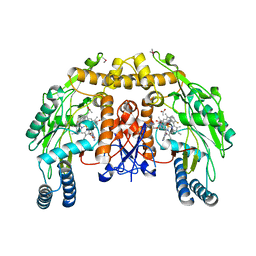 | | Structure of the endothelial nitric oxide synthase heme domain in complex with 6-{[(3R,4R)-4-(2-{[(2R/2S)-1-(3-fluorophenyl)propan-2-yl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(2R)-1-(3-fluorophenyl)propan-2-yl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2011-04-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cyclopropyl- and methyl-containing inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 21, 2013
|
|
7QKE
 
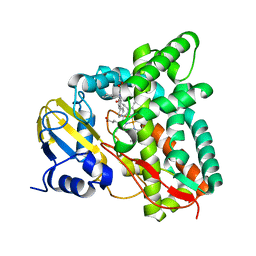 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with inhibitor (surface entropy reduction mutant) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase, ethyl 1-(cyclohexylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | | Authors: | Snee, M, Tunnicliffe, R, Leys, D, Levy, C, Katariya, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7QNN
 
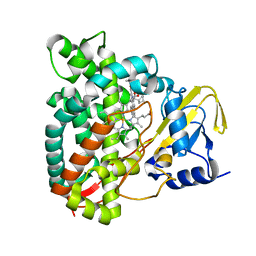 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with inhibitor (surface entropy reduction mutant) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase, ethyl 1-(cyclopentylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | | Authors: | Snee, M, Tunnicliffe, R, Leys, D, Levy, C, Katariya, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7QRA
 
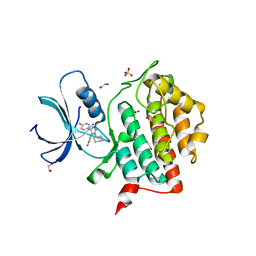 | | Crystal structure of CK1 delta in complex with VN725 | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-cyclohexyl-5-(4-fluorophenyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Nemec, V, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3SRS
 
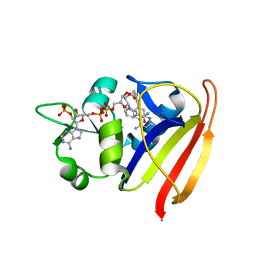 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 7-(5-bromo-2-ethoxyphenyl)-6-methylquinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SU3
 
 | | Crystal structure of NS3/4A protease in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SUG
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SU4
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, NS3 protease,NS4A protein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
7QR9
 
 | | Crystal structure of CK1 delta in complex with PK-09-82 | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(4-fluorophenyl)-3-(pyridin-4-ylmethyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7QRB
 
 | | Crystal structure of CK1 delta in complex with PK-09-129 | | Descriptor: | 3-(dimethylamino)-~{N}-[4-[4-(4-fluorophenyl)-5-(1~{H}-pyrrolo[2,3-b]pyridin-4-yl)imidazol-1-yl]cyclohexyl]propane-1-sulfonamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3SV8
 
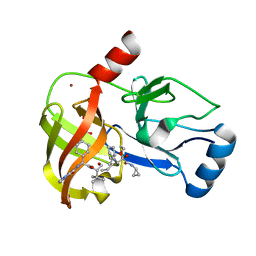 | | Crystal structure of NS3/4A protease variant D168A in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
6TNC
 
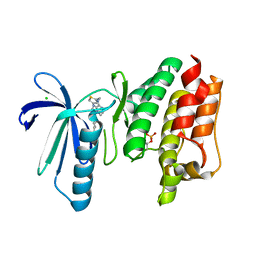 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 46 | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, N-cyclopropyl-4-{8-[(thiophen-2-ylmethyl)amino]imidazo[1,2-a]pyrazin-3-yl}benzamide | | Authors: | Marquardt, T, Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
6TOH
 
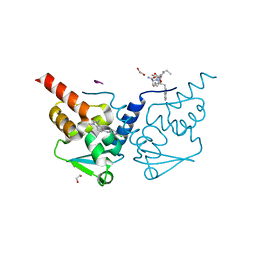 | | Crystal structure of human BCL6 BTB domain in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[(1,3-dimethyl-2-oxidanylidene-benzimidazol-5-yl)amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Shetty, K, Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
7QVK
 
 | | NM-02 in complex with HER2-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NM-02, Receptor tyrosine-protein kinase erbB-2, ... | | Authors: | Cowan, R, Hall, G, Carr, M. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Co-crystallisation and humanisation of an anti-HER2 single-domain antibody as a theranostic tool.
Plos One, 18, 2023
|
|
3T3C
 
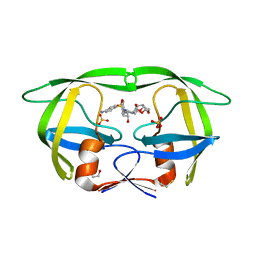 | | Structure of HIV PR resistant patient derived mutant (comprising 22 mutations) in complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, BETA-MERCAPTOETHANOL, HIV-1 protease, ... | | Authors: | Rezacova, P, Kozisek, M, Konvalinka, J, Saskova, K.G. | | Deposit date: | 2011-07-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations in HIV-1 gag and pol Compensate for the Loss of Viral Fitness Caused by a Highly Mutated Protease.
Antimicrob.Agents Chemother., 56, 2012
|
|
6T1Y
 
 | | Cryo-EM structure of phalloidin-stabilized F-actin (copolymerized) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Merino, F, Raunser, S. | | Deposit date: | 2019-10-07 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments.
Structure, 28, 2020
|
|
7QVJ
 
 | | ESTROGEN RECEPTOR ALPHA IN COMPLEX WITH COMPOUND 29 | | Descriptor: | 2,2-bis(fluoranyl)-3-[(1~{R},3~{R})-1-[6-fluoranyl-3-[2-(3-fluoranylpropylamino)ethoxy]-2-methyl-phenyl]-3-methyl-1,3,4,9-tetrahydropyrido[3,4-b]indol-2-yl]propan-1-ol, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Zwitterionic Series of Selective Estrogen Receptor Degrader-Antagonists.
J.Med.Chem., 66, 2023
|
|
7QVL
 
 | | OESTROGEN RECEPTOR LIGAND BINDING DOMAIN IN COMPLEX WITH COMPOUND 38 | | Descriptor: | (2~{R})-3-[(1~{R},3~{R})-1-[5-fluoranyl-2-[2-(3-fluoranylpropylamino)ethoxy]-3-methyl-pyridin-4-yl]-3-methyl-1,3,4,9-tetrahydropyrido[3,4-b]indol-2-yl]-2-methyl-propanoic acid, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Zwitterionic Series of Selective Estrogen Receptor Degrader-Antagonists.
J.Med.Chem., 66, 2023
|
|
7QWN
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
6T6P
 
 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) at 1.57 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, GLYCEROL, PHOSPHATE ION | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
