3E84
 
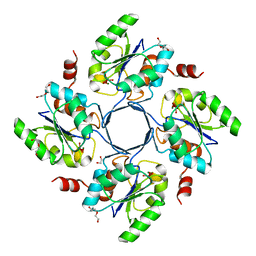 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
194D
 
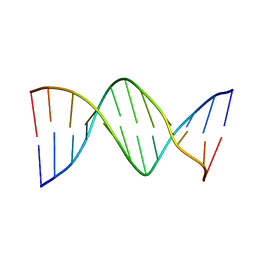 | | X-RAY STRUCTURES OF THE B-DNA DODECAMER D(CGCGTTAACGCG) WITH AN INVERTED CENTRAL TETRANUCLEOTIDE AND ITS NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*AP*AP*CP*GP*CP*G)-3') | | Authors: | Balendiran, K, Rao, S.T, Sekharudu, C.Y, Zon, G, Sundaralingam, M. | | Deposit date: | 1994-10-04 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1F4P
 
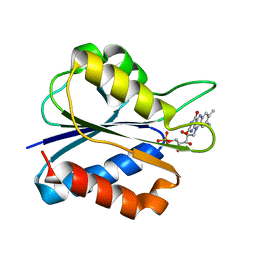 | | Y98W FLAVODOXIN MUTANT 1.5A (D. VULGARIS) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Reynolds, R.A, Watt, W, Watenpaugh, K.D. | | Deposit date: | 2000-06-08 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures and comparison of the Y98H (2.0 A) and Y98W (1.5 A) mutants of flavodoxin (Desulfovibrio vulgaris).
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1LJX
 
 | | THE STRUCTURE OF D(TPGPCPGPCPA)2 AT 293K: COMPARISON OF THE EFFECT OF SEQUENCE AND TEMPERATURE | | Descriptor: | 5'-D(*TP*GP*CP*GP*CP*A)-3', MAGNESIUM ION | | Authors: | Thiyagarajan, S, Satheesh Kumar, P, Rajan, S.S, Gautham, N. | | Deposit date: | 2002-04-23 | | Release date: | 2002-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of d(TGCGCA)2 at 293 K: comparison of the effects of sequence and temperature.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7JWW
 
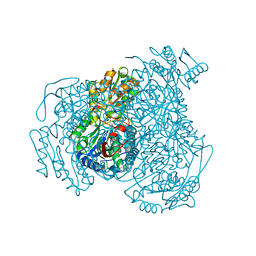 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 5-{4-[(Z)-2-hydroxyethenyl]phenyl}-1-methyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7TPU
 
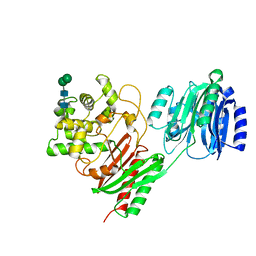 | | Crystal structure of a chitinase-modifying protein from Fusarium vanettenii (Fvan-cmp) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-lactamase domain-containing protein, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Dowling, N.V, Naumann, T.A, Price, N.P.J, Rose, D.R. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of a polyglycine hydrolase determined using a RoseTTAFold model.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1W2Y
 
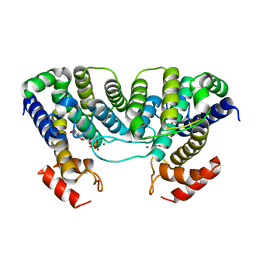 | | The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue dUpNHp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Galperin, M.Y, Vagin, A.A, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-07-09 | | Release date: | 2004-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of a Complex of Campylobacter Jejuni Dutpase with Substrate Analogue Sheds Light on the Mechanism and Suggests the "Basic Module" for Dimeric D(C/U)Tpases
J.Mol.Biol., 342, 2004
|
|
6R2J
 
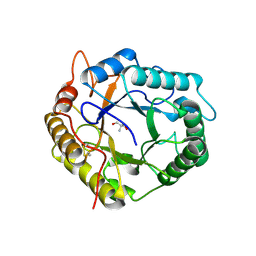 | | Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2019-03-18 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7BK3
 
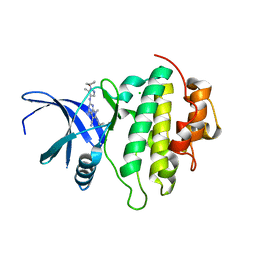 | | Crystal structure of CHK1-10pt-mutant complex with compound 45 | | Descriptor: | 4-amino-7-methyl-2-({5-methyl-1-[(3S)-oxolan-3-yl]-1H-pyrazol-4-yl}amino)-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJX
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 26 | | Descriptor: | 4-amino-7-methyl-2-[(1-methyl-1H-pyrazol-4-yl)amino]-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJJ
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 9 | | Descriptor: | 1H-pyrazolo[3,4-d]pyrimidin-4-amine, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BK1
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 32 | | Descriptor: | 4-amino-2-[(1,3-dimethyl-1H-pyrazol-4-yl)amino]-7-methyl-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BK2
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 44 | | Descriptor: | 4-amino-7-methyl-2-({5-methyl-1-[(3R)-oxolan-3-yl]-1H-pyrazol-4-yl}amino)-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7JWU
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 1-methyl-5-phenyl-6-{[(1R)-1-(pyridin-2-yl)ethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JWS
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 1-methyl-5-phenyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JWV
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 5-[4-(hydroxymethyl)phenyl]-1-methyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
8IDN
 
 | | Cryo-EM structure of RBD/E77-Fab complex | | Descriptor: | E77 Fab heavy chain, E77 Fab light chain, Spike protein S1, ... | | Authors: | Lu, D.F, Zhang, Z.C. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structure of the RBD-E77 Fab complex reveals neutralization and immune escape of SARS-CoV-2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7JWT
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 6-{[(1R)-1-(3-hydroxyphenyl)ethyl]sulfanyl}-1-methyl-5-phenyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
1D78
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | Descriptor: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | Authors: | Im, D, Shimamura, T, Iwata, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
5FHJ
 
 | |
4ORY
 
 | | Three-dimensional structure of the C65A-K59A double mutant of Human lipocalin-type Prostaglandin D Synthase holo, second crystal form | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4G1Z
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
5XHW
 
 | | Crystal structure of HddC from Yersinia pseudotuberculosis | | Descriptor: | Putative 6-deoxy-D-mannoheptose pathway protein, SULFATE ION | | Authors: | Park, J, Kim, H, Kim, S, Shin, D.H. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-glycero-alpha-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis.
Biochim. Biophys. Acta, 1866, 2018
|
|
4G2H
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
