2Q0M
 
 | | Tricarbonylmanganese(I)-lysozyme complex : a structurally characterized organometallic protein | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Razavet, M, Artero, V, Cavazza, C, Oudart, Y, Fontecilla-Camps, J.C, Fontecave, M. | | Deposit date: | 2007-05-22 | | Release date: | 2007-12-11 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tricarbonylmanganese(I)-lysozyme complex: a structurally characterized organometallic protein.
Chem.Commun.(Camb.), 2007
|
|
4APT
 
 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
2PW8
 
 | | Crystal structure of sulfo-hirudin complexed to thrombin | | Descriptor: | Hirudin variant-1, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Liu, C.C, Brustad, E, Liu, W, Schultz, P.G. | | Deposit date: | 2007-05-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin.
J.Am.Chem.Soc., 129, 2007
|
|
3AGB
 
 | |
3AHH
 
 | | H142A mutant of Phosphoketolase from Bifidobacterium Breve complexed with acetyl thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-ACETYL-THIAMINE DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
2OLN
 
 | | NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: closed form at 1.15 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PYRIDINE-2-CARBOXYLIC ACID, SODIUM ION, ... | | Authors: | Carrell, C.J, Bruckner, R.C, Venci, D, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2007-01-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: structures of closed and open forms at 1.15 and 1.90 A resolution
Structure, 15, 2007
|
|
3AG2
 
 | | Bovine Heart Cytochrome c Oxidase in the Carbon Monoxide-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2PA0
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sugahara, M, Tanaka, Y, Matsuura, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
2OPG
 
 | | The crystal structure of the 10th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein, SODIUM ION | | Authors: | Gileadi, C, Phillips, C, Elkins, J, Papagrigoriou, E, Ugochukwu, E, Gorrec, F, Savitsky, P, Umeano, C, Berridge, G, Gileadi, O, Salah, E, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 10th PDZ domain of MPDZ
To be Published
|
|
3C0V
 
 | | Crystal structure of cytokinin-specific binding protein in complex with cytokinin and Ta6Br12 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytokinin-specific binding protein, ... | | Authors: | Pasternak, O, Bujacz, A, Biesiadka, J, Bujacz, G, Sikorski, M, Jaskolski, M. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MAD phasing using the (Ta(6)Br(12))(2+) cluster: a retrospective study
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2P0O
 
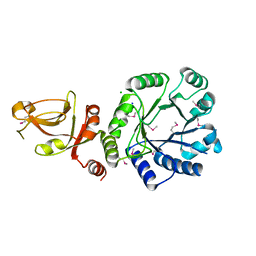 | | Crystal structure of a conserved protein from locus EF_2437 in Enterococcus faecalis with an unknown function | | Descriptor: | CHLORIDE ION, Hypothetical protein DUF871, SODIUM ION | | Authors: | Cuff, M.E, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a conserved protein from locus EF_2437 in Enterococcus faecalis with an unknown function.
TO BE PUBLISHED
|
|
4A1R
 
 | | The Structure of Serratia marcescens Lip, a membrane bound component of the Type VI Secretion System. | | Descriptor: | 1,2-ETHANEDIOL, LIP, SODIUM ION | | Authors: | Rao, V.A, Shepherd, S.M, English, G, Coulthurst, S.J, Hunter, W.N. | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Structure of Serratia Marcescens Lip, a Membrane-Bound Component of the Type Vi Secretion System
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2Q95
 
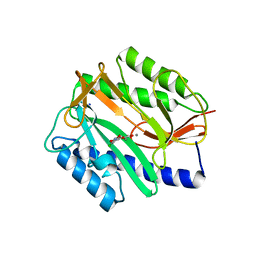 | | E. coli methionine aminopeptidase Mn-form with inhibitor A05 | | Descriptor: | 5-(2-CHLORO-4-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q92
 
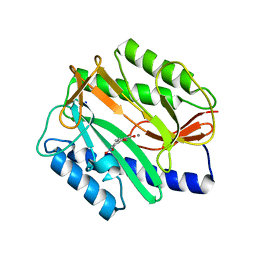 | | E. coli methionine aminopeptidase Mn-form with inhibitor B23 | | Descriptor: | 5-(2-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
4ARU
 
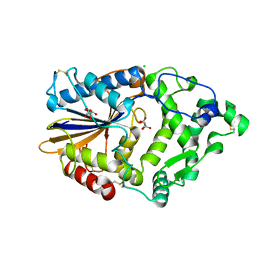 | | Hafnia Alvei phytase in complex with tartrate | | Descriptor: | CHLORIDE ION, HISTIDINE ACID PHOSPHATASE, L(+)-TARTARIC ACID, ... | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
3AVP
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with Pantothenol | | Descriptor: | (2S)-2,4-dihydroxy-N-(3-hydroxypropyl)-3,3-dimethylbutanamide, CITRATE ANION, GLYCEROL, ... | | Authors: | Chetnani, B, Kumar, P, Abhinav, K.V, Chhibber, M, Surolia, A, Vijayan, M. | | Deposit date: | 2011-03-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Location and conformation of pantothenate and its derivatives in Mycobacterium tuberculosis pantothenate kinase: insights into enzyme action
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1H88
 
 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | Descriptor: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-29 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
2QA0
 
 | | Structure of Adeno-Associated virus serotype 8 | | Descriptor: | Capsid protein, SODIUM ION | | Authors: | Nam, H.-J. | | Deposit date: | 2007-06-14 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of adeno-associated virus serotype 8, a gene therapy vector.
J.Virol., 81, 2007
|
|
4B52
 
 | | Crystal structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa | | Descriptor: | BACILLOLYSIN, CALCIUM ION, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Ruf, A, Stihle, M, Benz, J, Schmidt, M, Sobek, H. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of Gentlyase, the Neutral Metalloprotease of Paenibacillus Polymyxa
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4B2J
 
 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | CHLORIDE ION, DODECIN, MAGNESIUM ION, ... | | Authors: | Staudt, H, Hoesl, M, Dreuw, A, Serdjukow, S, Oesterhelt, D, Budisa, N, Wachtveitl, J, Grininger, M. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed Manipulation of a Flavoprotein Photocycle.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2NXB
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, SODIUM ION | | Authors: | Filippakopoulos, P, Bullock, A, Cooper, C, Keates, K, Berridge, G, Pike, A, Bunkoczi, G, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3A66
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N/D370Y mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOATE-DIMER HYDROLASE, 6-AMINOHEXANOIC ACID, ... | | Authors: | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2009-08-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
3AGA
 
 | | Crystal structure of RCC-bound red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | 3-{(2Z,3S,4S)-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-2-[(5R)-2-[(3-ethyl-5-formyl-4-methyl-1H-pyrrol-2-yl)methyl]-5-(methoxycarbonyl)-3-methyl-4-oxo-4,5-dihydrocyclopenta[b]pyrrol-6(1H)-ylidene]-4-methyl-3,4-dihydro-2H-pyrrol-3-yl}propanoic acid, Red chlorophyll catabolite reductase, chloroplastic, ... | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the substrate-bound forms of red chlorophyll catabolite reductase: implications for site-specific and stereospecific reaction
J.Mol.Biol., 402, 2010
|
|
1QY2
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | 2-ISOPROPYL-3-METHOXYPYRAZINE, CADMIUM ION, Major Urinary Protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
3AKR
 
 | |
