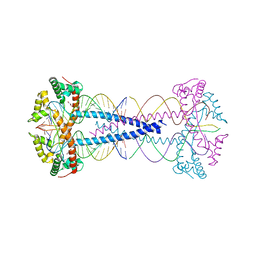
7ZIE
 
 | |
2KH1
 
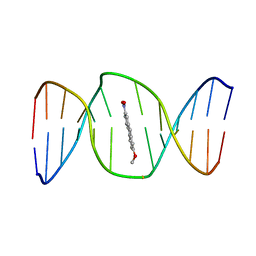 | | 2-Hydroxy-7-nitrofluorene covalently linked into a 13mer DNA duplex - solution structure of the face-up orientation | | Descriptor: | 5'-D(*CP*GP*AP*CP*GP*TP*(3DR)P*TP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*AP*(3DR)P*AP*CP*GP*TP*CP*G)-3', 7-nitro-9H-fluoren-2-ol | | Authors: | Dallmann, A, Pfaffe, M, Muegge, C, Mahrwald, R, Kovalenko, S.A, Ernsting, N.P. | | Deposit date: | 2009-03-23 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Local THz Time Domain Spectroscopy of Duplex DNA via Fluorescence of an Embedded Probe.
J.Phys.Chem.B, 113, 2009
|
|
7V0C
 
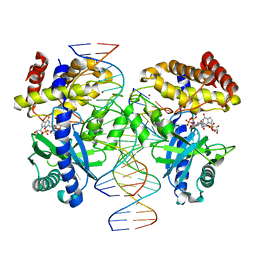 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, MANGANESE (II) ION, Palindromic DNA18, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG
To Be Published
|
|
7V0R
 
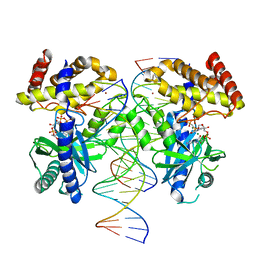 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -ppcpG(2 ,5 )pA | | Descriptor: | Cyclic GMP-AMP synthase, MAGNESIUM ION, Palindromic DNA18, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -ppcpG(2 ,5 )pA
To Be Published
|
|
2KH7
 
 | |
2KH0
 
 | | 2-Hydroxy-7-nitrofluorene covalently linked into a 13mer DNA duplex - solution structure of the face-down orientation | | Descriptor: | 5'-D(*CP*GP*AP*CP*GP*TP*(3DR)P*TP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*AP*(3DR)P*AP*CP*GP*TP*CP*G)-3', 7-nitro-9H-fluoren-2-ol | | Authors: | Dallmann, A, Pfaffe, M, Muegge, C, Mahrwald, R, Kovalenko, S.A, Ernsting, N.P. | | Deposit date: | 2009-03-23 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Local THz Time Domain Spectroscopy of Duplex DNA via Fluorescence of an Embedded Probe
J.Phys.Chem.B, 113, 2009
|
|
2KH8
 
 | |
8ECC
 
 | |
1YFI
 
 | | Crystal Structure of restriction endonuclease MspI in complex with its cognate DNA in P212121 space group | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', Type II restriction enzyme MspI | | Authors: | Xu, Q.S, Kucera, R.B, Roberts, R.J, Guo, H.-C. | | Deposit date: | 2005-01-02 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure.
Protein Sci., 14, 2005
|
|
2KHU
 
 | | Solution Structure of the Ubiquitin-Binding Motif of Human Polymerase Iota | | Descriptor: | Immunoglobulin G-binding protein G, DNA polymerase iota | | Authors: | Bomar, M.G, D'Souza, S, Bienko, M, Dikic, I, Walker, G. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unconventional Ubiquitin Recognition by the Ubiquitin-Binding Motif within the Y Family DNA Polymerases iota and Rev1.
Mol.Cell, 37, 2010
|
|
6F8J
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F96
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(4-methoxyphenyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-[(4-methoxyphenyl)amino]-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
1XA2
 
 | |
1XAM
 
 | | Cobalt hexammine induced tautameric shift in Z-DNA: structure of d(CGCGCA).d(TGCGCG) in two crystal forms. | | Descriptor: | CGCGCA, COBALT HEXAMMINE(III), TG, ... | | Authors: | Thiyagarajan, S, Rajan, S.S, Gautham, N. | | Deposit date: | 2004-08-26 | | Release date: | 2004-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Cobalt hexammine induced tautomeric shift in Z-DNA: the structure of d(CGCGCA)*d(TGCGCG) in two crystal forms.
Nucleic Acids Res., 32, 2004
|
|
6F86
 
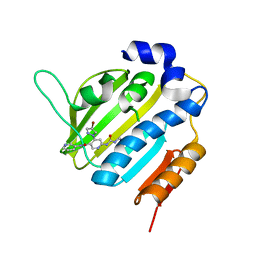 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F94
 
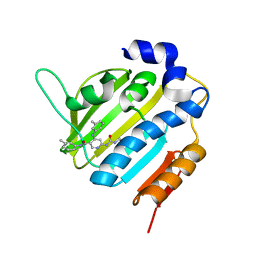 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(3-methyphenyl)amino]-N-(3-methyphenyl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-~{N}-(3-methylphenyl)-4-[(3-methylphenyl)amino]pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6G8S
 
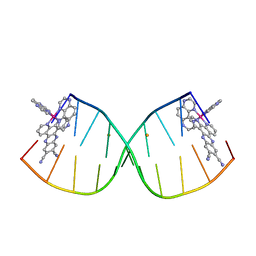 | | [Ru(TAP)2(11,12-CN2-dppz)]2+ bound to d(CCGGACCCGG/CCGGGTCCGG)2 | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*CP*GP*GP*AP*CP*CP*CP*GP*G)-3'), DNA (5'-D(*CP*CP*GP*GP*GP*TP*CP*CP*GP*G)-3'), ... | | Authors: | McQuaid, K.T, Hall, J.P, Cardin, C.J. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | X-ray Crystal Structures Show DNA Stacking Advantage of Terminal Nitrile Substitution in Ru-dppz Complexes.
Chemistry, 24, 2018
|
|
1ZR4
 
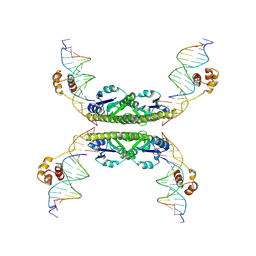 | | Structure of a Synaptic gamma-delta Resolvase Tetramer Covalently linked to two Cleaved DNAs | | Descriptor: | AAA, TCAGTGTCCGATAATTTAT, TTATCGGACACTG, ... | | Authors: | Li, W, Kamtekar, S, Xiong, Y, Sarkis, G.J, Grindley, N.D, Steitz, T.A. | | Deposit date: | 2005-05-19 | | Release date: | 2005-08-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs.
Science, 309, 2005
|
|
1ZR2
 
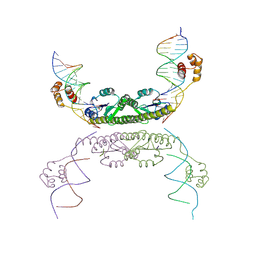 | | Structure of a Synaptic gamma-delta Resolvase Tetramer Covalently Linked to two Cleaved DNAs | | Descriptor: | AAA, TCAGTGTCCGATAATTTAT, TTATCGGACACTG, ... | | Authors: | Li, W, Kamtekar, S, Xiong, Y, Sarkis, G.J, Grindley, N.D, Steitz, T.A. | | Deposit date: | 2005-05-18 | | Release date: | 2005-08-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a synaptic gamma delta resolvase tetramer covalently linked to two cleaved DNAs.
Science, 309, 2005
|
|
258D
 
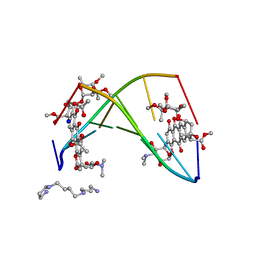 | | FACTORS AFFECTING SEQUENCE SELECTIVITY ON NOGALAMYCIN INTERCALATION: THE CRYSTAL STRUCTURE OF D(TGTACA)-NOGALAMYCIN | | Descriptor: | ACETATE ION, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3'), NOGALAMYCIN, ... | | Authors: | Smith, C.K, Brannigan, J.A, Moore, M.H. | | Deposit date: | 1996-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Factors affecting DNA sequence selectivity of nogalamycin intercalation: the crystal structure of d(TGTACA)2-nogalamycin2.
J.Mol.Biol., 263, 1996
|
|
7ZBG
 
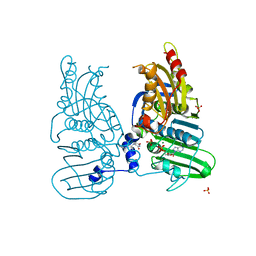 | | Human Topoisomerase II Beta ATPase ADP | | Descriptor: | 4-[(2S,3R)-3-[3,5-bis(oxidanylidene)piperazin-1-ium-1-yl]butan-2-yl]piperazin-4-ium-2,6-dione, ADENOSINE-5'-DIPHOSPHATE, DNA topoisomerase 2-beta, ... | | Authors: | Ling, E.M, Basle, A, Cowell, I.G, Blower, T.R, Austin, C.A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A comprehensive structural analysis of the ATPase domain of human DNA topoisomerase II beta bound to AMPPNP, ADP, and the bisdioxopiperazine, ICRF193.
Structure, 30, 2022
|
|
8ALQ
 
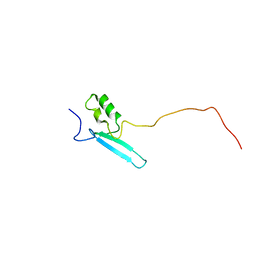 | |
2KYY
 
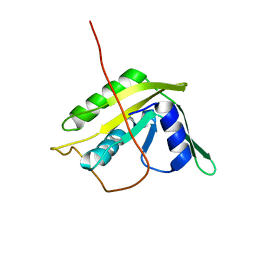 | | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea, Northeast Structural Genomics Consortium Target NeR70A | | Descriptor: | Possible ATP-dependent DNA helicase RecG-related protein | | Authors: | Eletsky, A, Mills, J.L, Lee, H, Maglaqui, M, Ciccosanti, C, Hamilton, K, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea
To be Published
|
|
1Z1B
 
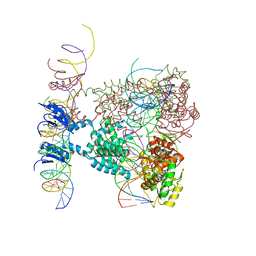 | | Crystal structure of a lambda integrase dimer bound to a COC' core site | | Descriptor: | 26-MER DNA, 29-MER DNA, 5'-D(*CP*T*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
1Z63
 
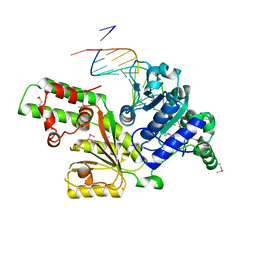 | | Sulfolobus solfataricus SWI2/SNF2 ATPase core in complex with dsDNA | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*A*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*AP*AP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*GP*TP*CP*TP*TP*CP*GP*GP*CP*AP*AP*TP*TP*TP*TP*TP*T)-3', Helicase of the snf2/rad54 family | | Authors: | Duerr, H, Koerner, C, Mueller, M, Hickmann, V, Hopfner, K.P. | | Deposit date: | 2005-03-21 | | Release date: | 2005-05-17 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA.
Cell(Cambridge,Mass.), 121, 2005
|
|
