9B5T
 
 | |
9C0I
 
 | |
9C0B
 
 | | E.coli GroEL + PBZ1587 inhibitor | | Descriptor: | 60 kDa chaperonin, N-(2-{4-[4-(aminomethyl)benzene-1-sulfonamido]phenyl}-1,3-benzoxazol-5-yl)-5-chlorothiophene-2-sulfonamide | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2024-05-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Bis-sulfonamido-2-phenylbenzoxazoles Validate the GroES/EL Chaperone System as a Viable Antibiotic Target.
J.Am.Chem.Soc., 146, 2024
|
|
9B57
 
 | |
9B5K
 
 | |
9CVE
 
 | |
9B55
 
 | |
9CVF
 
 | |
9B5F
 
 | |
9EO4
 
 | | Outward-open structure of human dopamine transporter bound to cocaine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nielsen, J.C, Salomon, K, Kalenderoglou, I.E, Bargmeyer, S, Pape, T, Shahsavar, A, Loland, C.J. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human dopamine transporter in complex with cocaine.
Nature, 632, 2024
|
|
9B5S
 
 | |
9B5B
 
 | |
9C0C
 
 | | E.coli GroEL apoenzyme | | Descriptor: | 60 kDa chaperonin | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2024-05-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Bis-sulfonamido-2-phenylbenzoxazoles Validate the GroES/EL Chaperone System as a Viable Antibiotic Target.
J.Am.Chem.Soc., 146, 2024
|
|
9B5M
 
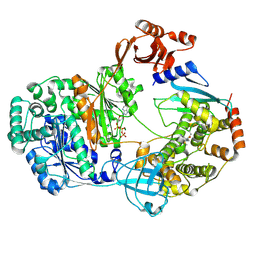 | |
9CSI
 
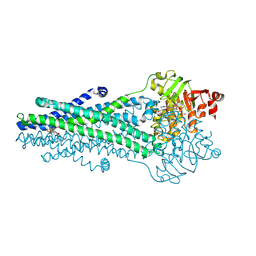 | | A. baumannii MsbA Bound to Cerastecin Compound 5 | | Descriptor: | 3,3'-[(1,4-dioxobutane-1,4-diyl)bis(azanediyl)]bis[(4-butylbenzene-1-sulfonamido)benzoic acid], Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION, ... | | Authors: | Klein, D.J, Ishchenko, A, Soisson, S, Cheng, R, Hennig, M. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
9C0D
 
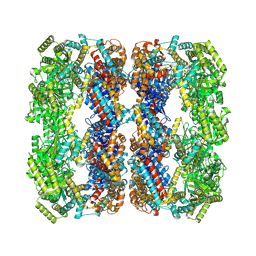 | | E.Faecium GroEL | | Descriptor: | Chaperonin GroEL | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2024-05-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Bis-sulfonamido-2-phenylbenzoxazoles Validate the GroES/EL Chaperone System as a Viable Antibiotic Target.
J.Am.Chem.Soc., 146, 2024
|
|
9B5C
 
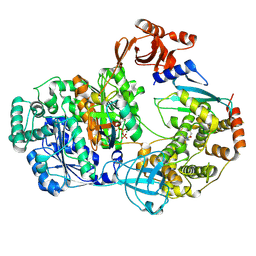 | |
9LYZ
 
 | |
9B5Q
 
 | |
9B5O
 
 | |
9B5N
 
 | |
9B33
 
 | |
9B5U
 
 | |
9C4M
 
 | | Crystal Structure of A. baumannii GuaB dCBS with inhibitor G6 (3826) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(morpholin-4-yl)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Differential effects of inosine monophosphate dehydrogenase (IMPDH/GuaB) inhibition in Acinetobacter baumannii and Escherichia coli.
J.Bacteriol., 2024
|
|
9F9Y
 
 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 2024
|
|
