1TR1
 
 | | CRYSTAL STRUCTURE OF E96K MUTATED BETA-GLUCOSIDASE A FROM BACILLUS POLYMYXA, AN ENZYME WITH INCREASED THERMORESISTANCE | | Descriptor: | BETA-GLUCOSIDASE A, GLYCEROL | | Authors: | Sanz-Aparicio, J, Hermoso, J.A, Martinez-Ripoll, M, Gonzalez-Perez, B, Polaina, J. | | Deposit date: | 1998-03-12 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
1ICJ
 
 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | Descriptor: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1BRZ
 
 | | SOLUTION STRUCTURE OF THE SWEET PROTEIN BRAZZEIN, NMR, 43 STRUCTURES | | Descriptor: | BRAZZEIN | | Authors: | Caldwell, J.E, Abildgaard, F, Dzakula, Z, Ming, D, Hellekant, G, Markley, J.L. | | Deposit date: | 1998-03-12 | | Release date: | 1998-07-01 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the thermostable sweet-tasting protein brazzein.
Nat.Struct.Biol., 5, 1998
|
|
1CBU
 
 | | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE (COBU) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE, SULFATE ION | | Authors: | Thompson, T.B, Thomas, M.G, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 1998-03-12 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase from Salmonella typhimurium determined to 2.3 A resolution,.
Biochemistry, 37, 1998
|
|
1A7C
 
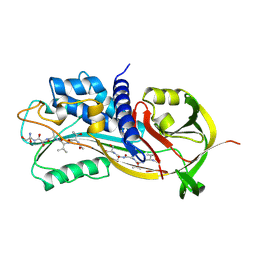 | | HUMAN PLASMINOGEN ACTIVATOR INHIBITOR TYPE-1 IN COMPLEX WITH A PENTAPEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-ribopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PENTAPEPTIDE, ... | | Authors: | Xue, Y, Inghardt, T, Sjolin, L, Deinum, J. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interfering with the inhibitory mechanism of serpins: crystal structure of a complex formed between cleaved plasminogen activator inhibitor type 1 and a reactive-centre loop peptide
Structure, 6, 1998
|
|
1A7D
 
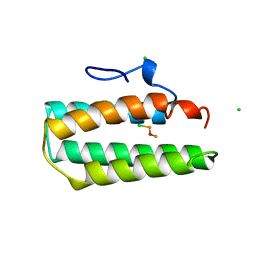 | |
1A7G
 
 | |
1IAO
 
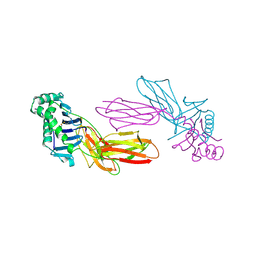 | | CLASS II MHC I-AD IN COMPLEX WITH OVALBUMIN PEPTIDE 323-339 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-AD | | Authors: | Scott, C.A, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 1998-03-13 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues.
Immunity, 8, 1998
|
|
1A7H
 
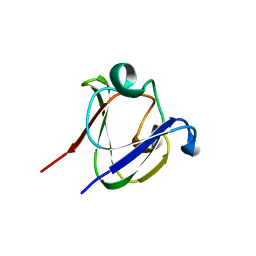 | | GAMMA S CRYSTALLIN C-TERMINAL DOMAIN | | Descriptor: | GAMMAS CRYSTALLIN | | Authors: | Basak, A.K, Slingsby, C. | | Deposit date: | 1998-03-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The C-terminal domains of gammaS-crystallin pair about a distorted twofold axis.
Protein Eng., 11, 1998
|
|
2GCC
 
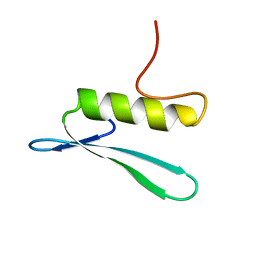 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
2IAD
 
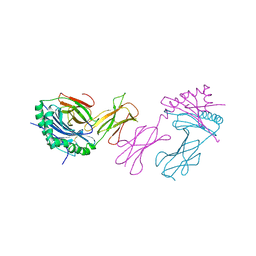 | |
2ERC
 
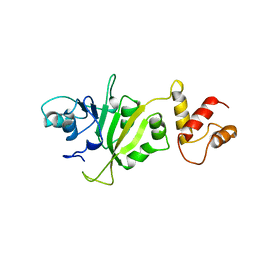 | | CRYSTAL STRUCTURE OF ERMC' A RRNA-METHYL TRANSFERASE | | Descriptor: | RRNA METHYL TRANSFERASE | | Authors: | Bussiere, D.E, Muchmore, S.W, Dealwis, C.G, Schluckebier, G, Abad-Zapatero, C. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of ErmC', an rRNA methyltransferase which mediates antibiotic resistance in bacteria.
Biochemistry, 37, 1998
|
|
8OHM
 
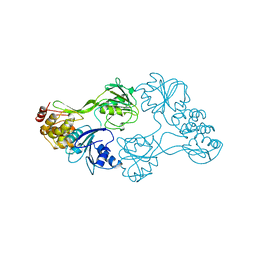 | |
3GCC
 
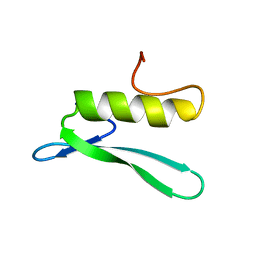 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, 46 STRUCTURES | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
1GCC
 
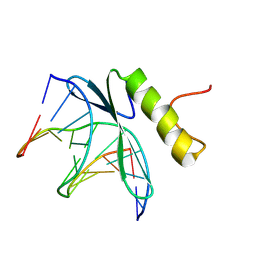 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF GCC-BOX BINDING DOMAIN OF ATERF1 AND GCC-BOX DNA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*GP*CP*GP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*CP*GP*CP*CP*AP*GP*C)-3'), ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 | | Authors: | Yamasaki, K, Allen, M.D, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
1TBC
 
 | |
1TAC
 
 | |
12E8
 
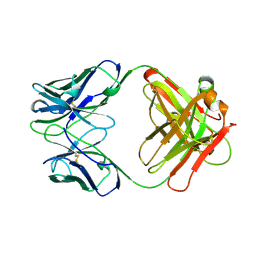 | | 2E8 FAB FRAGMENT | | Descriptor: | IGG1-KAPPA 2E8 FAB (HEAVY CHAIN), IGG1-KAPPA 2E8 FAB (LIGHT CHAIN) | | Authors: | Rupp, B, Trakhanov, S. | | Deposit date: | 1998-03-14 | | Release date: | 1998-08-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a monoclonal 2E8 Fab antibody fragment specific for the low-density lipoprotein-receptor binding region of apolipoprotein E refined at 1.9 A.
Acta Crystallogr.,Sect.D, null, 1999
|
|
1A7I
 
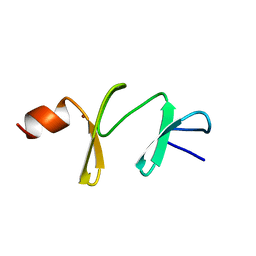 | | AMINO-TERMINAL LIM DOMAIN FROM QUAIL CYSTEINE AND GLYCINE-RICH PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | QCRP2 (LIM1), ZINC ION | | Authors: | Kontaxis, G, Konrat, R, Kraeutler, B, Weiskirchen, R, Bister, K. | | Deposit date: | 1998-03-15 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and intramodular dynamics of the amino-terminal LIM domain from quail cysteine- and glycine-rich protein CRP2.
Biochemistry, 37, 1998
|
|
1LDP
 
 | |
1A7N
 
 | |
1A7M
 
 | | LEUKAEMIA INHIBITORY FACTOR CHIMERA (MH35-LIF), NMR, 20 STRUCTURES | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Hinds, M.G, Maurer, T, Zhang, J.-G, Nicola, N.A, Norton, R.S. | | Deposit date: | 1998-03-16 | | Release date: | 1999-04-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of leukemia inhibitory factor.
J.Biol.Chem., 273, 1998
|
|
1PGJ
 
 | | X-RAY STRUCTURE OF 6-PHOSPHOGLUCONATE DEHYDROGENASE FROM THE PROTOZOAN PARASITE T. BRUCEI | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, SULFATE ION | | Authors: | Dohnalek, J, Phillips, C, Gover, S, Barrett, M.P, Adams, M.J. | | Deposit date: | 1998-03-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | A 2.8 A resolution structure of 6-phosphogluconate dehydrogenase from the protozoan parasite Trypanosoma brucei: comparison with the sheep enzyme accounts for differences in activity with coenzyme and substrate analogues.
J.Mol.Biol., 282, 1998
|
|
1A7L
 
 | | DOMINANT B-CELL EPITOPE FROM THE PRES2 REGION OF HEPATITIS B VIRUS IN THE FORM OF AN INSERTED PEPTIDE SEGMENT IN MALTODEXTRIN-BINDING PROTEIN | | Descriptor: | MALE-B363, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Saul, F.A, Vulliez-Lenormand, B, Lema, F, Bentley, G.A. | | Deposit date: | 1998-03-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a dominant B-cell epitope from the preS2 region of hepatitis B virus in the form of an inserted peptide segment in maltodextrin-binding protein.
J.Mol.Biol., 280, 1998
|
|
1A7R
 
 | |
