6SFI
 
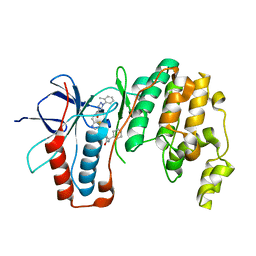 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6S9Y
 
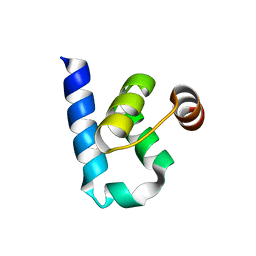 | |
6SFJ
 
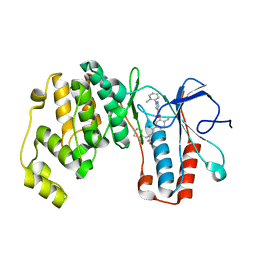 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6QE1
 
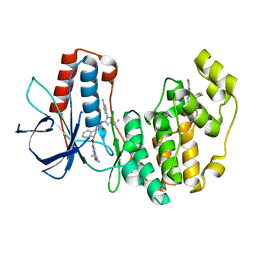 | | P38 alpha complex with AR117046 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{R})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | P38 alpha complex with AR117045 and AR117046
To Be Published
|
|
6QL5
 
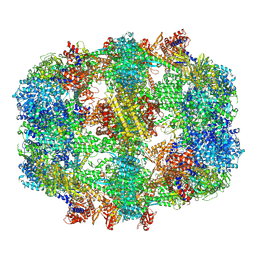 | | Structure of fatty acid synthase complex with bound gamma subunit from Saccharomyces cerevisiae at 2.8 angstrom | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
6RD8
 
 | | CryoEM structure of Polytomella F-ATP synthase, c-ring position 2, focussed refinement of Fo and peripheral stalk | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6SFO
 
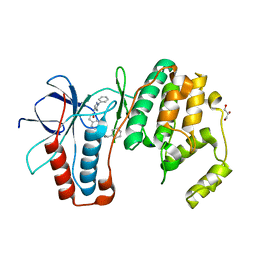 | | MAPK14 with bound inhibitor SR-318 | | Descriptor: | 5-azanyl-~{N}-[[4-(3-cyclohexylpropylcarbamoyl)phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Schroeder, M, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MAPK14 with bound inhibitor SR-318
To Be Published
|
|
8B6G
 
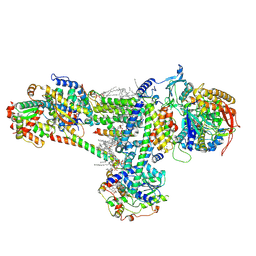 | | Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6J
 
 | | Cryo-EM structure of cytochrome bc1 complex (complex-III) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apocytochrome b, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8BOM
 
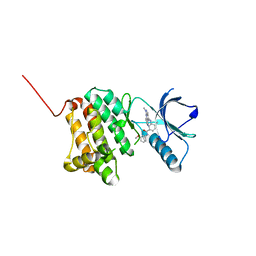 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 14 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(3-methoxyphenyl)-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
6SM1
 
 | | Wild type immunoglobulin light chain (WT-1) | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Immunoglobulin lambda variable 2-14, ... | | Authors: | Kazman, P, Vielberg, M.-T, Cendales, M.D.P, Hunziger, L, Weber, B, Hegenbart, U, Zacharias, M, Koehler, R, Schoenland, S, Groll, M, Buchner, J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fatal amyloid formation in a patient's antibody light chain is caused by a single point mutation.
Elife, 9, 2020
|
|
6SFK
 
 | | Crystal structure of p38 alpha in complex with compound 81 (MCP42) | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
8BLO
 
 | | Human Urea Transporter UT-A (N-Terminal Domain Model) | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Urea transporter 2, di-heneicosanoyl phosphatidyl choline | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Scacioc, A, Wang, D, McKinley, G, Fernandez-Cid, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Burgess-Brown, N.A, van Putte, W, Duerr, K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
8BWU
 
 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Transcription factor ETV6,Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Konkolova, E, Klima, M, Boura, E, Jin, J, Kaniskan, H.U, Han, Y, Vedadi, M. | | Deposit date: | 2022-12-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Application of established computational techniques to identify potential SARS-CoV-2 Nsp14-MTase inhibitors in low data regimes
Digit Discov, 2024
|
|
7ZWT
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-chlorophenyl)amino]-~{N}-(pyridin-2-ylmethyl)-1,3-thiazole-4-carboxamide, B-cell lymphoma 6 protein, ... | | Authors: | Gunnell, E.A, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
8CF9
 
 | | Crystal structure of the human PXR ligand-binding domain in complex with sclareol | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 1 group I member 2, sclareol | | Authors: | Carivenc, C, Derosa, Q, Grimaldi, M, Boulahtouf, A, Balaguer, P, Bourguet, W. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human PXR ligand-binding domain in complex with sclareol
To Be Published
|
|
6QDZ
 
 | | P38 alpha complex with AR117045 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{S})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | P38 alpha complex with AR117045
To Be Published
|
|
7Z38
 
 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7Z37
 
 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
6TCA
 
 | | Phosphorylated p38 and MAPKAPK2 complex with inhibitor | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14, N-[5-(dimethylsulfamoyl)-2-methylphenyl]-1-phenyl-5-propyl-1H-pyrazole-4-carboxamide | | Authors: | Sok, P, Remenyi, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | MAP Kinase-Mediated Activation of RSK1 and MK2 Substrate Kinases.
Structure, 28, 2020
|
|
1BAL
 
 | |
1FZ0
 
 | |
1BF3
 
 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND ARG 42 REPLACED BY LYS (R42K), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lys42 and Ser42 variants of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens reveal that Arg42 is essential for NADPH binding.
Eur.J.Biochem., 253, 1998
|
|
1FZI
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM I PRESSURIZED WITH XENON GAS | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ALPHA CHAIN, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
1AZH
 
 | |
