8C9T
 
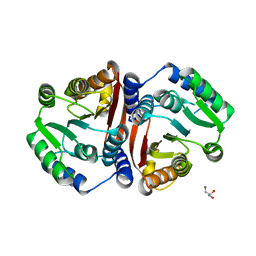 | | Catechol O-methyltransferase from Streptomyces avermitilis | | Descriptor: | GLYCEROL, Putative O-methyltransferase | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization and Extended Substrate Scope Analysis of Two Mg 2+ -Dependent O-Methyltransferases from Bacteria.
Chembiochem, 24, 2023
|
|
7SGZ
 
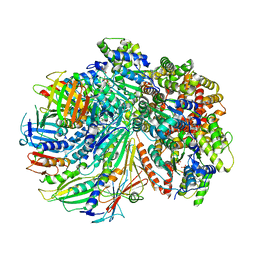 | | Structure of the yeast Rad24-RFC loader bound to DNA and the closed 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6WJV
 
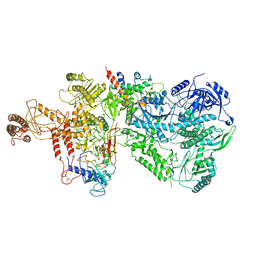 | | Structure of the Saccharomyces cerevisiae polymerase epsilon holoenzyme | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit B, DNA polymerase epsilon subunit C, ... | | Authors: | Yuan, Z, Georgescu, R, Schauer, G.D, O'Donnell, M, Li, H. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the polymerase epsilon holoenzyme and atomic model of the leading strand replisome.
Nat Commun, 11, 2020
|
|
8C9S
 
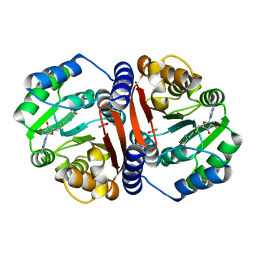 | |
7Q0P
 
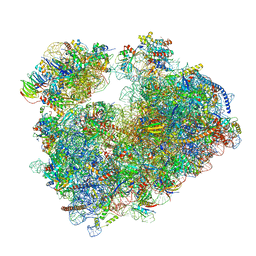 | | Structure of the Candida albicans 80S ribosome in complex with anisomycin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
8D6K
 
 | | Sco GlgEI-V279S in complex with cyclohexyl carbasugar | | Descriptor: | (1R,4S,5S,6R)-4-(cyclohexylamino)-5,6-dihydroxy-2-(hydroxymethyl)cyclohex-2-en-1-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, DI(HYDROXYETHYL)ETHER | | Authors: | Jayasinghe, T.J, Ronning, D.R. | | Deposit date: | 2022-06-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Synthesis of C 7 /C 8 -cyclitols and C 7 N-aminocyclitols from maltose and X-ray crystal structure of Streptomyces coelicolor GlgEI V279S in a complex with an amylostatin GXG-like derivative.
Front Chem, 10, 2022
|
|
6OH7
 
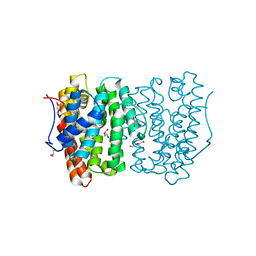 | |
7SH2
 
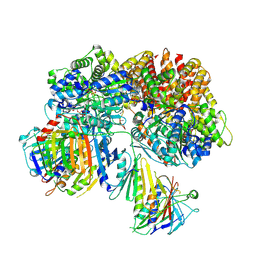 | | Structure of the yeast Rad24-RFC loader bound to DNA and the open 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6OH8
 
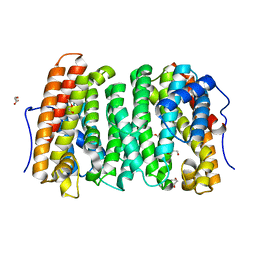 | |
8QPY
 
 | |
8QSX
 
 | |
7KDY
 
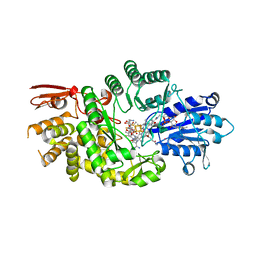 | | Crystal structure of Streptomyces tokunonesis TokK with hydroxycobalamin, 5'-deoxyadenosine, methionine, and (2R)-pantetheinylated carbapenam | | Descriptor: | (2R,3R,5R)-3-{[2-({N-[(2R)-2,4-dihydroxy-3,3-dimethylbutanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-7-oxo-1-azabicyclo[3.2.0]heptane-2-carboxylic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Knox, H.L, Booker, S.J, Boal, A.K. | | Deposit date: | 2020-10-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Structure of a B 12 -dependent radical SAM enzyme in carbapenem biosynthesis.
Nature, 602, 2022
|
|
7KDX
 
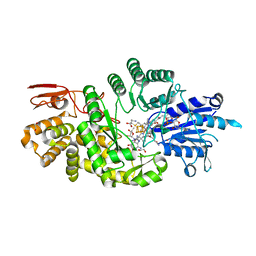 | | Crystal structure of Streptomyces tokunonesis TokK with hydroxycobalamin, 5'-deoxyadenosine, and methionine | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | Authors: | Knox, H.L, Booker, S.J, Boal, A.K. | | Deposit date: | 2020-10-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure of a B 12 -dependent radical SAM enzyme in carbapenem biosynthesis.
Nature, 602, 2022
|
|
6V8P
 
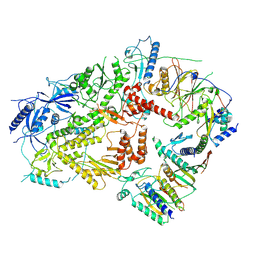 | | Structure of DNA Polymerase Zeta (Apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Malik, R, Gomez-Llorente, Y, Ubarretxena-Belandia, I, Aggarwal, A.K. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8WDG
 
 | |
6LZ1
 
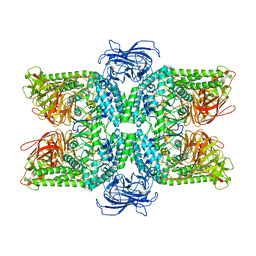 | | Structure of S.pombe alpha-mannosidase Ams1 | | Descriptor: | Ams1, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of fission yeast tetrameric alpha-mannosidase Ams1.
Febs Open Bio, 10, 2020
|
|
8WJW
 
 | |
8WK9
 
 | |
8WK5
 
 | |
8WK7
 
 | |
8WKA
 
 | |
8GYH
 
 | | Crystal structure of Fic25 (apo form) from Streptomyces ficellus | | Descriptor: | DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, IMIDAZOLE | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8GYI
 
 | | Crystal structure of Fic25 (holo form) from Streptomyces ficellus | | Descriptor: | DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8WU7
 
 | | Structure of a cis-Geranylfarnesyl Diphosphate Synthase from Streptomyces clavuligerus | | Descriptor: | Isoprenyl transferase | | Authors: | Li, F.R, Wang, Q.L, Pan, X.M, Dong, L.B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery, Structure, and Engineering of a cis-Geranylfarnesyl Diphosphate Synthase.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8GYJ
 
 | | Crystal structure of Fic25 complexed with PLP-(5S,6S)-N2-acetyl-DADH adduct from Streptomyces ficellus | | Descriptor: | (2~{S},5~{S},6~{S})-2-acetamido-6-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-5,7-bis(oxidanyl)heptanoic acid, DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
