1GJP
 
 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 4-OXOSEBACIC ACID | | Descriptor: | 4-OXODECANEDIOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1GJQ
 
 | | Pseudomonas aeruginosa cd1 nitrite reductase reduced cyanide complex | | Descriptor: | CYANIDE ION, HEME C, HEME D, ... | | Authors: | Nurizzo, D, Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanide Binding to Cd(1) Nitrite Reductase from Pseudomonas Aeruginosa: Role of the Active-Site His369 in Ligand Stabilization.
Biochem.Biophys.Res.Commun., 291, 2002
|
|
1GJR
 
 | | Ferredoxin-NADP+ Reductase complexed with NADP+ by COCRYSTALLIZATION | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-08-01 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Coenzyme Recognition and Binding Revealed by Crystal Structure Analysis of Ferredoxin-Nadp(+) Reductase Complexed with Nadp(+)
J.Mol.Biol., 319, 2002
|
|
1GJS
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GJU
 
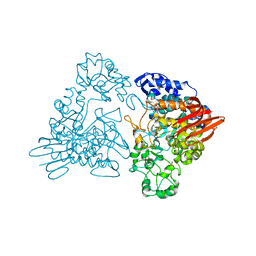 | | Maltosyltransferase from Thermotoga maritima | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-02 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
1GJV
 
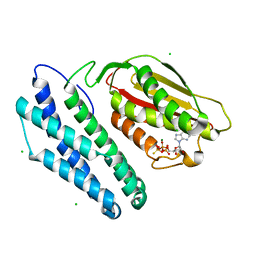 | | Branched-chain alpha-ketoacid dehydrogenase kinase (BCK) complxed with ATP-gamma-S | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Machius, M, Chuang, J.L, Wynn, M.R, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Rat Bckd Kinase: Nucleotide-Induced Domain Communication in a Mitochondrial Protein Kinase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GJW
 
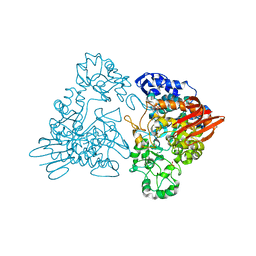 | | Thermotoga maritima maltosyltransferase complex with maltose | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION, alpha-D-glucopyranose, ... | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
1GJX
 
 | | Solution structure of the lipoyl domain of the chimeric dihydrolipoyl dehydrogenase P64K from Neisseria meningitidis | | Descriptor: | PYRUVATE DEHYDROGENASE | | Authors: | Tozawa, K, Broadhurst, R.W, Raine, A.R.C, Fuller, C, Alvarez, A, Guillen, G, Padron, G, Perham, R.N. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Lipoyl Domain of the Chimeric Dihydrolipoyl Dehydrogenase P64K from Neisseria Meningitidis
Eur.J.Biochem., 268, 2001
|
|
1GJY
 
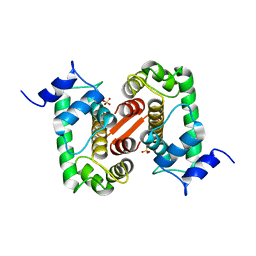 | | The X-ray structure of the Sorcin Calcium Binding Domain (SCBD) provides insight into the phosphorylation and calcium dependent processess | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Ilari, A, Johnson, K.A, Nastopoulos, V, Tsernoglou, D, Chiancone, E. | | Deposit date: | 2001-08-06 | | Release date: | 2002-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Sorcin Calcium Binding Domain Provides a Model of Ca(2+)-Dependent Processes in the Full-Length Protein
J.Mol.Biol., 317, 2002
|
|
1GJZ
 
 | |
1GK0
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOSPORIN ACYLASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1GK1
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | CEPHALOSPORIN ACYLASE, GLYCEROL | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1GK2
 
 | |
1GK3
 
 | |
1GK4
 
 | | HUMAN VIMENTIN COIL 2B FRAGMENT (CYS2) | | Descriptor: | ACETATE ION, VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
1GK5
 
 | | Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor | | Descriptor: | Pro-epidermal growth factor,Protransforming growth factor alpha | | Authors: | Chamberlin, S.G, Brennan, L, Puddicombe, S.M, Davies, D.E, Turner, D.L. | | Deposit date: | 2001-08-08 | | Release date: | 2002-08-08 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Megf/Tgfalpha44-50 Chimeric Growth Factor.
Eur.J.Biochem., 268, 2001
|
|
1GK6
 
 | | Human vimentin coil 2B fragment linked to GCN4 leucine zipper (Z2B) | | Descriptor: | VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
1GK7
 
 | | HUMAN VIMENTIN COIL 1A FRAGMENT (1A) | | Descriptor: | SULFATE ION, VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
1GK8
 
 | | Rubisco from Chlamydomonas reinhardtii | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Taylor, T.C. | | Deposit date: | 2001-08-09 | | Release date: | 2001-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | First Crystal Structure of Rubisco from a Green Alga, Chlamydomonas Reinhardtii.
J.Biol.Chem., 276, 2001
|
|
1GK9
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GKA
 
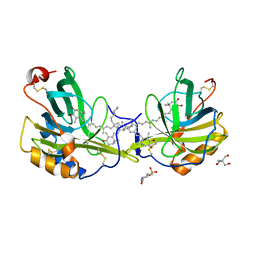 | | The molecular basis of the coloration mechanism in lobster shell. beta-crustacyanin at 3.2 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASTAXANTHIN, ... | | Authors: | Cianci, M, Rizkallah, P.J, Olczak, A, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2001-08-10 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | The Molecular Basis of the Coloration Mechanism in Lobster Shell: Beta -Crustacyanin at 3.2-A Resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GKB
 
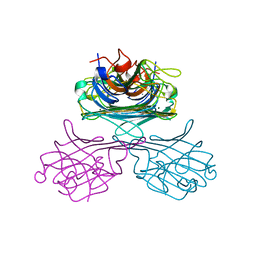 | | CONCANAVALIN A, NEW CRYSTAL FORM | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MAGNESIUM ION, ... | | Authors: | Kantardjieff, K, Rupp, B, Hoechtl, P, Segelke, B. | | Deposit date: | 2001-08-10 | | Release date: | 2001-08-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Concanavalin a in a Dimeric Crystal Form: Revisiting Structural Accuracy and Molecular Flexibility
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GKC
 
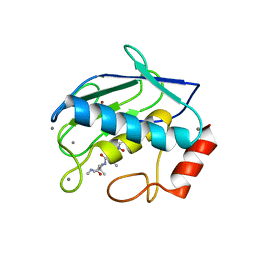 | | MMP9-inhibitor complex | | Descriptor: | 92 KDA TYPE IV COLLAGENASE, CALCIUM ION, N~2~-[(2R)-2-{[formyl(hydroxy)amino]methyl}-4-methylpentanoyl]-N,3-dimethyl-L-valinamide, ... | | Authors: | Rowsell, S, Pauptit, R.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Mmp9 in Complex with a Reverse Hydroxamate Inhibitor
J.Mol.Biol., 319, 2002
|
|
1GKD
 
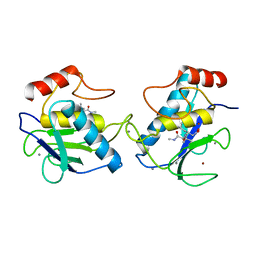 | | MMP9 active site mutant-inhibitor complex | | Descriptor: | 2-AMINO-N,3,3-TRIMETHYLBUTANAMIDE, 2-{[FORMYL(HYDROXY)AMINO]METHYL}-4-METHYLPENTANOIC ACID, 92 KDA TYPE IV COLLAGENASE, ... | | Authors: | Rowsell, S, Pauptit, R.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Mmp9 in Complex with a Reverse Hydroxamate Inhibitor
J.Mol.Biol., 319, 2002
|
|
