1S7O
 
 | | Crystal structure of putative DNA binding protein SP_1288 from Streptococcus pygenes | | Descriptor: | Hypothetical UPF0122 protein SPy1201/SpyM3_0842/SPs1042/spyM18_1152 | | Authors: | Oganesyan, V, Pufan, R, DeGiovanni, A, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-29 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the putative DNA-binding protein SP_1288 from Streptococcus pyogenes.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7Z3Y
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013545 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)-2,6-bis(fluoranyl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Scaletti, E.R, Stenmark, P. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013545
To Be Published
|
|
7Z5B
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013546 | | Descriptor: | 2-[4-(2,4-dimethyl-1~{H}-imidazol-5-yl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013546
To Be Published
|
|
7Z5R
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012035 | | Descriptor: | (7~{R})-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)bicyclo[4.2.0]octa-1,3,5-triene-7-carboxamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012035
To Be Published
|
|
7ZC7
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941
To Be Published
|
|
2IVK
 
 | | Crystal structure of the periplasmic endonuclease Vvn complexed with a 16-bp DNA | | Descriptor: | 5'-D(*AP*AP*TP*TP*CP*GP*AP*TP*CP*GP *AP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*TP*CP*GP*AP*TP*CP *GP*AP*AP*TP*T)-3', 5'-D(*GP*AP*AP*TP*TP*CP*GP*AP*TP*CP *GP*AP*AP*TP*TP*C)-3', ... | | Authors: | Wang, Y.T, Yang, W.J, Li, C.L, Doudeva, L.G, Yuan, H.S. | | Deposit date: | 2006-06-14 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Sequence-Dependent DNA Cleavage by Nonspecific Endonucleases.
Nucleic Acids Res., 35, 2007
|
|
8CEX
 
 | |
8CEY
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH11233 | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-(3,3-dimethylcyclobutyl)imidazo[5,1-b][1,3]thiazole-3-carboxamide | | Authors: | Kosenina, S, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH11233
To Be Published
|
|
4FNC
 
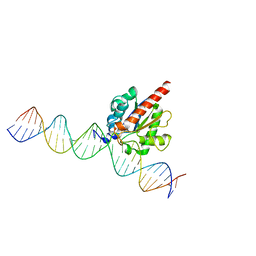 | | Human TDG in a post-reactive complex with 5-hydroxymethyluracil (5hmU) | | Descriptor: | 5-HYDROXYMETHYL URACIL, DNA (28-MER), DNA (29-MER), ... | | Authors: | Hashimoto, H, Hong, S, Bhagwat, A.S, Zhang, X, Cheng, X. | | Deposit date: | 2012-06-19 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
6C2F
 
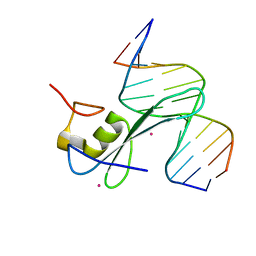 | | MBD2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-08 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MBD2 in complex with methylated DNA
to be published
|
|
5Z68
 
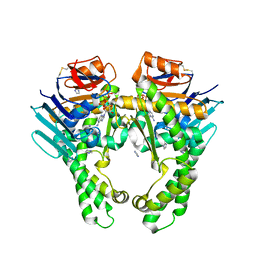 | | Structure of the recombination mediator protein RecF-ATP in RecFOR pathway | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA replication and repair protein RecF, IMIDAZOLE, ... | | Authors: | Tang, Q, Liu, Y.-P, Yan, X.-X. | | Deposit date: | 2018-01-22 | | Release date: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATP-dependent conformational change in ABC-ATPase RecF serves as a switch in DNA repair.
Sci Rep, 8, 2018
|
|
7ZG3
 
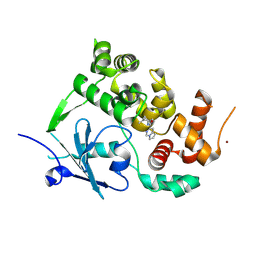 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228 | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-[(1~{S})-1,2,2-trimethylcyclopropyl]pyrrolo[1,2-c]pyrimidine-3-carboxamide | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-04-01 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228
To Be Published
|
|
7U38
 
 | | Pixantrone tethered DNA duplex | | Descriptor: | 1,2-ETHANEDIOL, Pixantrone AP conjugate-modified DNA | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2022-02-26 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Characterization of a Covalent Conjugate between Pixantrone and an Abasic Site in DNA
To Be Published
|
|
8APL
 
 | | Vaccinia virus DNA helicase D5 residues 323-785 hexamer with bound DNA processed in C6 | | Descriptor: | Primase D5 | | Authors: | Burmeister, W.P, Hutin, S, Ling, W.L, Grimm, C, Schoehn, G. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Vaccinia Virus DNA Helicase Structure from Combined Single-Particle Cryo-Electron Microscopy and AlphaFold2 Prediction.
Viruses, 14, 2022
|
|
1D33
 
 | | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-RAY diffraction studies | | Descriptor: | 5'-D(*CP*GP*CP*(G49)P*CP*G)-3', DAUNOMYCIN, MAGNESIUM ION | | Authors: | Wang, A.H.-J, Gao, Y.-G, Liaw, Y.-C, Li, Y.-K. | | Deposit date: | 1991-02-27 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-ray diffraction studies.
Biochemistry, 30, 1991
|
|
4GLE
 
 | | SacUVDE in complex with 6-4PP-containing DNA | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*CP*AP*AP*GP*GP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*CP*CP*(64T)P*(5PY)P*GP*AP*CP*GP*AP*CP*G)-3', SULFATE ION, ... | | Authors: | Meulenbroek, E.M, Peron Cane, C, Jala, I, Iwai, S, Moolenaar, G.F, Goosen, N, Pannu, N.S. | | Deposit date: | 2012-08-14 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | UV damage endonuclease employs a novel dual-dinucleotide flipping mechanism to recognize different DNA lesions.
Nucleic Acids Res., 41, 2013
|
|
5JJV
 
 | |
2CQG
 
 | | Solution structure of the RNA binding domain of TAR DNA-binding protein-43 | | Descriptor: | TAR DNA-binding protein-43 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of TAR DNA-binding protein-43
To be Published
|
|
3SSD
 
 | | DNA binding domain of restriction endonuclease bound to DNA | | Descriptor: | 5-methylcytosine-specific restriction enzyme B, DNA (5'-D(*A*GP*CP*TP*AP*CP*CP*GP*GP*TP*CP*TP*C)-3'), DNA (5'-D(*T*GP*AP*GP*AP*(5CM)P*CP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Sukackaite, R, Grazulis, S, Siksnys, V. | | Deposit date: | 2011-07-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine.
Nucleic Acids Res., 40, 2012
|
|
8KE7
 
 | |
3FQB
 
 | |
1M6I
 
 | | Crystal Structure of Apoptosis Inducing Factor (AIF) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Programmed cell death protein 8 | | Authors: | Ye, H, Cande, C, Stephanou, N.C, Jiang, S, Gurbuxani, S, Larochette, N, Daugas, E, Garrido, C, Kroemer, G, Wu, H. | | Deposit date: | 2002-07-16 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA binding is required for the apoptogenic action of apoptosis inducing factor.
Nat.Struct.Biol., 9, 2002
|
|
1P8K
 
 | |
2NQ4
 
 | | Solution Structures of a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3' | | Authors: | Bhattacharyya, D, Wu, Y, Chaney, S, Campbell, S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a DNA Dodecamer Duplex with and without a Cisplatin 1,2-d(GG) Intrastrand Cross-Link: Comparison with the Same DNA Duplex Containing an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link
Biochemistry, 46, 2007
|
|
2NQ0
 
 | | Solution Structures of a DNA Dodecamer Duplex with a Cisplatin 1,2-d(GG) Intrastrand Cross-Link | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', Cisplatin | | Authors: | Wu, Y, Bhattacharyya, D, Chaney, S, Campbell, S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a DNA Dodecamer Duplex with and without a Cisplatin 1,2-d(GG) Intrastrand Cross-Link: Comparison with the Same DNA Duplex Containing an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link
Biochemistry, 46, 2007
|
|
