1P4S
 
 | |
2M3C
 
 | | Solution Structure of gammaM7-Crystallin | | Descriptor: | Crystallin, gamma M7 | | Authors: | Mahler, B, Wu, Z. | | Deposit date: | 2013-01-16 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Fish Eye Lens Protein, gamma M7-Crystallin.
Biochemistry, 52, 2013
|
|
2MLW
 
 | | New Cyt-like delta-endotoxins from Dickeya dadantii - CytC protein | | Descriptor: | Type-1Ba cytolytic delta-endotoxin | | Authors: | Loth, K, Costechareyre, D, Effantin, G, Rahbe, Y, Condemine, G, Landon, C, Da Silva, P. | | Deposit date: | 2014-03-05 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | New Cyt-like delta-endotoxins from Dickeya dadantii: structure and aphicidal activity.
Sci Rep, 5, 2015
|
|
1PBZ
 
 | |
2M16
 
 | | P75/LEDGF PWWP Domain | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Crowe, B.L, Foster, M.P. | | Deposit date: | 2012-11-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes.
Nucleic Acids Res., 41, 2013
|
|
2M7G
 
 | |
2LY8
 
 | | The budding yeast chaperone Scm3 recognizes the partially unfolded dimer of the centromere-specific Cse4/H4 histone variant | | Descriptor: | Budding yeast chaperone Scm3 | | Authors: | Hong, J, Feng, H, Zhou, Z, Ghirlando, R, Bai, Y. | | Deposit date: | 2012-09-13 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of Functionally Conserved Regions in the Structure of the Chaperone/CenH3/H4 Complex.
J.Mol.Biol., 425, 2013
|
|
1NE3
 
 | | Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744 | | Descriptor: | 30S ribosomal protein S28E | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J.R, Ramelot, T.A, Kennedy, M, Edwards, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum.
Protein Sci., 12, 2003
|
|
1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
1OMT
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (STANDARD NOESY ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
2MRN
 
 | |
2MZP
 
 | | Structure and dynamics of the acidosis-resistant a162H mutant of the switch region of troponin I bound to the regulatory domain of troponin C | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Pineda Sanabria, S.E, Robertson, I.M, Sykes, B.D. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Acidosis-Resistant A162H Mutant of the Switch Region of Troponin I Bound to the Regulatory Domain of Troponin C.
Biochemistry, 54, 2015
|
|
1Q2K
 
 | | Solution structure of BmBKTx1 a new potassium channel blocker from the Chinese Scorpion Buthus martensi Karsch | | Descriptor: | Neurotoxin BmK37 | | Authors: | Cai, Z, Xu, C, Xu, Y, Lu, W, Chi, C.W, Shi, Y, Wu, J. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmBKTx1, a New BK(Ca)(1) Channel Blocker from the Chinese Scorpion Buthus martensi Karsch(,).
Biochemistry, 43, 2004
|
|
1POZ
 
 | | SOLUTION STRUCTURE OF THE HYALURONAN BINDING DOMAIN OF HUMAN CD44 | | Descriptor: | CD44 antigen | | Authors: | Teriete, P, Banerji, S, Blundell, C.D, Kahmann, J.D, Pickford, A.R, Wright, A.J, Campbell, I.D, Jackson, D.G, Day, A.J. | | Deposit date: | 2003-06-16 | | Release date: | 2004-03-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Regulatory Hyaluronan Binding Domain in the Inflammatory Leukocyte Homing Receptor CD44.
Mol.Cell, 13, 2004
|
|
1OF9
 
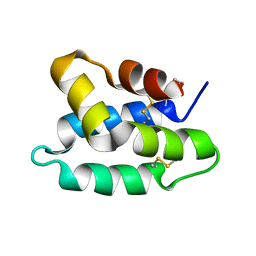 | | Solution structure of the pore forming toxin of entamoeba histolytica (Amoebapore A) | | Descriptor: | PORE-FORMING PEPTIDE AMEOBAPORE A | | Authors: | Hecht, O, Schleinkofer, K, Bruhn, H, Leippe, M, Van Nuland, N, Dingley, A.J, Grotzinger, J. | | Deposit date: | 2003-04-09 | | Release date: | 2004-02-26 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pore-Forming Protein of Entamoeba Histolytica
J.Biol.Chem., 279, 2004
|
|
1Q2T
 
 | | Solution structure of d(5mCCTCTCC)4 | | Descriptor: | 5'-D(*(MCY)P*CP*TP*CP*TP*CP*C)-3' | | Authors: | Leroy, J.-L. | | Deposit date: | 2003-07-26 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | T.T pair intercalation and duplex inter-conversion within i-motif tetramers
J.Mol.Biol., 333, 2003
|
|
1NOT
 
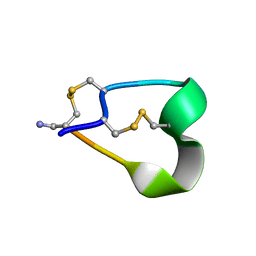 | | THE 1.2 ANGSTROM STRUCTURE OF G1 ALPHA CONOTOXIN | | Descriptor: | GI ALPHA CONOTOXIN | | Authors: | Guddat, L.W, Shan, L, Martin, J.L, Edmundson, A.B, Gray, W.R. | | Deposit date: | 1996-05-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Three-dimensional structure of the alpha-conotoxin GI at 1.2 A resolution
Biochemistry, 35, 1996
|
|
1P68
 
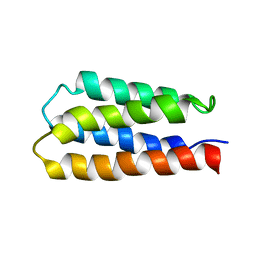 | | Solution structure of S-824, a de novo designed four helix bundle | | Descriptor: | De novo designed protein S-824 | | Authors: | Wei, Y, Kim, S, Fela, D, Baum, J, Hecht, M.H. | | Deposit date: | 2003-04-29 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a de novo protein from a designed combinatorial library.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1NRB
 
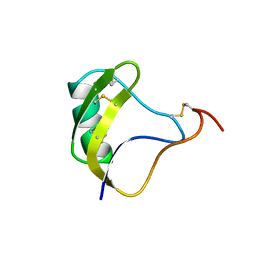 | |
1NRA
 
 | |
1NYD
 
 | | Solution structure of DNA quadruplex GCGGTGGAT | | Descriptor: | 5'-D(*GP*CP*GP*GP*TP*GP*GP*AP*T)-3' | | Authors: | Webba da Silva, M. | | Deposit date: | 2003-02-12 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Association of DNA quadruplexes through G:C:G:C tetrads. Solution structure of d(GCGGTGGAT).
Biochemistry, 42, 2003
|
|
1OAW
 
 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
1QLO
 
 | | Structure of the active domain of the herpes simplex virus protein ICP47 in water/sodium dodecyl sulfate solution determined by nuclear magnetic resonance spectroscopy | | Descriptor: | HERPES SIMPLEX VIRUS PROTEIN ICP47 | | Authors: | Pfaender, R, Neumann, L, Zweckstetter, M, Seger, C, Holak, T.A, Tampe, R. | | Deposit date: | 1999-09-09 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Active Domain of the Herpes Simplex Virus Protein Icp47 in Water/Sodium Dodecyl Sulfate Solution Determined by Nuclear Magnetic Resonance Spectroscopy.
Biochemistry, 38, 1999
|
|
1P6R
 
 | | Solution structure of the DNA binding domain of the repressor BlaI. | | Descriptor: | Penicillinase repressor | | Authors: | Melckebeke, H.V, Vreuls, C, Gans, P, Llabres, G, Filee, P, Joris, B, Simorre, J.P. | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structural study of BlaI: implications for the repression of genes involved in beta-lactam antibiotic resistance.
J.Mol.Biol., 333, 2003
|
|
1ON5
 
 | |
