4IHQ
 
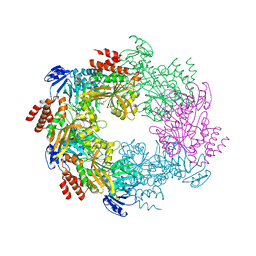 | | Archaellum Assembly ATPase FlaI bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, FlaI ATPase, ... | | Authors: | Reindl, S, Williams, G.J, Tainer, J.A. | | Deposit date: | 2012-12-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into FlaI Functions in Archaeal Motor Assembly and Motility from Structures, Conformations, and Genetics.
Mol.Cell, 49, 2013
|
|
1JLN
 
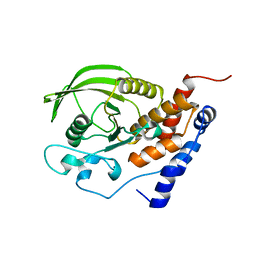 | | Crystal structure of the catalytic domain of protein tyrosine phosphatase PTP-SL/BR7 | | Descriptor: | Protein Tyrosine Phosphatase, receptor type, R | | Authors: | Szedlacsek, S.E, Aricescu, A.R, Fulga, T.A, Renault, L, Scheidig, A.J. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of PTP-SL/PTPBR7 catalytic domain: implications for MAP kinase regulation.
J.Mol.Biol., 311, 2001
|
|
4II7
 
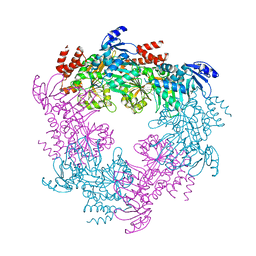 | | Archaellum Assembly ATPase FlaI | | Descriptor: | FlaI ATPase | | Authors: | Reindl, S, Williams, G.J, Tainer, J.A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Insights into FlaI Functions in Archaeal Motor Assembly and Motility from Structures, Conformations, and Genetics.
Mol.Cell, 49, 2013
|
|
3IT2
 
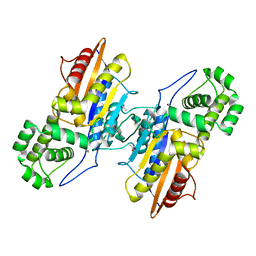 | | Crystal structure of ligand-free Francisella tularensis histidine acid phosphatase | | Descriptor: | ACETATE ION, Acid phosphatase | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
2CWW
 
 | | Crystal structure of Thermus thermophilus TTHA1280, a putative SAM-dependent RNA methyltransferase, in complex with S-adenosyl-L-homocysteine | | Descriptor: | ACETIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
8C6J
 
 | | Human spliceosomal PM5 C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Dybkov, O, Kastner, B, Luehrmann, R. | | Deposit date: | 2023-01-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Regulation of 3' splice site selection after step 1 of splicing by spliceosomal C* proteins.
Sci Adv, 9, 2023
|
|
8C0D
 
 | | UFL1/DDRGK1 bound to UFC1 | | Descriptor: | DDRGK domain-containing protein 1, E3 UFM1-protein ligase 1, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Magnussen, H.M, Kulathu, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8BZR
 
 | | UFC1-UFM1 conjugate | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-fold modifier 1, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Magnussen, H.M, Kulathu, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
1CZ1
 
 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS AT 1.85 A RESOLUTION | | Descriptor: | PROTEIN (EXO-B-(1,3)-GLUCANASE) | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-01-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
5IJ4
 
 | | Solution structure of AN1-type zinc finger domain from Cuz1 (Cdc48 associated ubiquitin-like/zinc-finger protein-1) | | Descriptor: | CDC48-associated ubiquitin-like/zinc finger protein 1, ZINC ION | | Authors: | Sun, Z.-Y.J, Hanna, J, Wagner, G, Bhanu, M.K, Allan, M, Arthanari, H. | | Deposit date: | 2016-03-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cuz1 AN1 Zinc Finger Domain: An Exposed LDFLP Motif Defines a Subfamily of AN1 Proteins.
Plos One, 11, 2016
|
|
1OAW
 
 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
1OEB
 
 | | Mona/Gads SH3C domain | | Descriptor: | CADMIUM ION, GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Harkiolaki, M, Lewitzky, M, Gilbert, R.J.C, Jones, E.Y, Bourette, R.P, Mouchiroud, G, Sondermann, H, Moarefi, I, Feller, S.M. | | Deposit date: | 2003-03-24 | | Release date: | 2003-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for SH3 Domain-Mediated High-Affinity Binding between Mona/Gads and Slp-76
Embo J., 22, 2003
|
|
7KZL
 
 | | Cyclopentane peptide nucleic acid in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*AP*TP*CP*AP*CP*AP*TP*C)-3'), IODIDE ION, ... | | Authors: | Botos, I, Appella, D.H. | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational constraints of cyclopentane peptide nucleic acids facilitate tunable binding to DNA.
Nucleic Acids Res., 49, 2021
|
|
1PVZ
 
 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
1QK1
 
 | | CRYSTAL STRUCTURE OF HUMAN UBIQUITOUS MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, UBIQUITOUS MITOCHONDRIAL, PHOSPHATE ION | | Authors: | Eder, M, Schlattner, U, Fritz-Wolf, K, Wallimann, T, Kabsch, W. | | Deposit date: | 1999-07-08 | | Release date: | 2000-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Ubiquitous Mitochondrial Creatine Kinase
Proteins: Struct.,Funct., Genet., 39, 2000
|
|
1PE4
 
 | | SOLUTION STRUCTURE OF TOXIN CN12 FROM CENTRUROIDES NOXIUS ALFA SCORPION TOXIN ACTING ON SODIUM CHANNELS. NMR STRUCTURE | | Descriptor: | Neurotoxin Cn11 | | Authors: | Del Rio-Portilla, F, Hernandez-Marin, E, Pimienta, G, Coronas, F.V, Zamudio, F.Z, Rodrguez de la Vega, R.C, Wanke, E, Possani, L.D. | | Deposit date: | 2003-05-21 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Cn12, a novel peptide from the Mexican scorpion Centruroides noxius with a typical beta-toxin sequence but with alpha-like physiological activity.
Eur.J.Biochem., 271, 2004
|
|
1NIX
 
 | |
1OAV
 
 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
7NOY
 
 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis in complex with substrate 1-hydroxyquinolin-4(1H)-one | | Descriptor: | 1-oxidanylquinolin-4-one, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
7NMK
 
 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis with bound methylation product 1-methoxyquinolin-4(1H)-one | | Descriptor: | 1-methoxy-4-oxoquinoline, 2-heptyl-1-hydroxyquinolin-4(1H)-one methyltransferase, FORMIC ACID, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.204 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
8OUJ
 
 | | Heterotrimeric Complex of Human ASCT2 with Syncytin-1 | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Syncytin-1 | | Authors: | Khare, S, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OUI
 
 | | Complex of ASCT2 with Suppressyn | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Suppressyn | | Authors: | Khare, S, Kumar, A, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OUH
 
 | | Complex of human ASCT2 with Syncytin-1 | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Syncytin-1 | | Authors: | Khare, S, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OUD
 
 | |
7NV1
 
 | | Human Pol Kappa holoenzyme with Ub-PCNA | | Descriptor: | DNA Primer, DNA Template, DNA polymerase kappa, ... | | Authors: | Lancey, C, De Biasio, A, Hamdan, S.M. | | Deposit date: | 2021-03-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Cryo-EM structure of human Pol kappa bound to DNA and mono-ubiquitylated PCNA.
Nat Commun, 12, 2021
|
|
