3O1Y
 
 | | Electron transfer complexes: Experimental mapping of the redox-dependent cytochrome c electrostatic surface | | Descriptor: | Cytochrome c, HEME C, NITRATE ION | | Authors: | De March, M, De Zorzi, R, Demitri, N, Gabbiani, C, Guerri, A, Casini, A, Messori, L, Geremia, S. | | Deposit date: | 2010-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nitrate as a probe of cytochrome c surface: crystallographic identification of crucial "hot spots" for protein-protein recognition.
J. Inorg. Biochem., 135, 2014
|
|
1W2O
 
 | | Deacetoxycephalosporin C synthase (with a N-terminal his-tag) in complex with Fe(II) and deacetoxycephalosporin C | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE, DEACETOXYCEPHALOSPORIN-C, FE (III) ION | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
2X08
 
 | | cytochrome c peroxidase: ascorbate bound to the engineered ascorbate binding site | | Descriptor: | ASCORBIC ACID, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Engineering the substrate specificity and reactivity of a heme protein: creation of an ascorbate binding site in cytochrome c peroxidase.
Biochemistry, 47, 2008
|
|
1GU6
 
 | | Structure of the Periplasmic Cytochrome c Nitrite Reductase from Escherichia coli | | Descriptor: | CALCIUM ION, CYTOCHROME C552, GLYCEROL, ... | | Authors: | Bamford, V.A, Angove, H.C, Seward, H.E, Thomson, A.J, Cole, J.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Spectroscopy of the Periplasmic Cytochrome C Nitrite Reductase from Escherichia Coli
Biochemistry, 41, 2002
|
|
2CCY
 
 | | STRUCTURE OF FERRICYTOCHROME C(PRIME) FROM RHODOSPIRILLUM MOLISCHIANUM AT 1.67 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Finzel, B.C, Weber, P.C, Hardman, K.D, Salemme, F.R. | | Deposit date: | 1985-08-27 | | Release date: | 1986-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of ferricytochrome c' from Rhodospirillum molischianum at 1.67 A resolution.
J.Mol.Biol., 186, 1985
|
|
8BJW
 
 | |
2UUN
 
 | | Crystal structure of C-phycocyanin from Phormidium, Lyngbya spp. (Marine) and Spirulina sp. (Fresh water) shows two different ways of energy transfer between two hexamers. | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN, PHYCOCYANOBILIN | | Authors: | Satyanarayana, L, Patel, A, Mishra, S, K Ghosh, P, Suresh, C.G. | | Deposit date: | 2007-03-05 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Phormidium, Lyngbya Spp. (Marine) and Spirulina Sp. (Fresh Water) Shows Two Different Ways of Energy Transfer between Two Hexamers.
To be Published
|
|
2GMX
 
 | | Selective Aminopyridine-Based C-Jun N-terminal Kinase inhibitors with cellular activity | | Descriptor: | C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, N-(4-AMINO-5-CYANO-6-ETHOXYPYRIDIN-2-YL)-2-(4-BROMO-2,5-DIMETHOXYPHENYL)ACETAMIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-04-07 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aminopyridine-Based c-Jun N-Terminal Kinase Inhibitors with Cellular Activity and Minimal Cross-Kinase Activity.
J.Med.Chem., 49, 2006
|
|
6YQG
 
 | | Crystal structure of native Phycocyanin in spacegroup P63 at 1.45 Angstroms. | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-17 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6YPQ
 
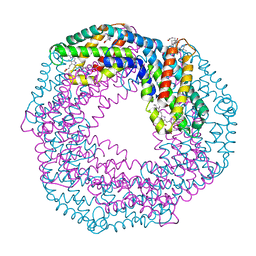 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup R32 at 1.29 Angstroms | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, GLYCINE, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2O0I
 
 | | crystal structure of the R185A mutant of the N-terminal domain of the Group B Streptococcus Alpha C protein | | Descriptor: | C protein alpha-antigen | | Authors: | Hogle, J.M, Filman, D.J, Baron, M.J, Madoff, L.C, Iglesias, A. | | Deposit date: | 2006-11-27 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of a glycosaminoglycan binding region of the alpha C protein that mediates entry of group B streptococci into host cells.
J.Biol.Chem., 282, 2007
|
|
1AB2
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE SRC HOMOLOGY 2 DOMAIN OF C-ABL | | Descriptor: | C-ABL TYROSINE KINASE SH2 DOMAIN | | Authors: | Overduin, M, Rios, C.B, Mayer, B.J, Baltimore, D, Cowburn, D. | | Deposit date: | 1993-07-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the src homology 2 domain of c-abl.
Cell(Cambridge,Mass.), 70, 1992
|
|
2V6H
 
 | | Crystal structure of the C1 domain of cardiac myosin binding protein-C | | Descriptor: | MYOSIN-BINDING PROTEIN C, CARDIAC-TYPE | | Authors: | Govata, L, Carpenter, L, Da Fonseca, P.C.A, Helliwell, J.R, Rizkallah, P.J, Flashman, E, Chayen, N.E, Redwood, C, Squire, J.M. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the C1 domain of cardiac myosin binding protein-C: implications for hypertrophic cardiomyopathy.
J. Mol. Biol., 378, 2008
|
|
1Y7Y
 
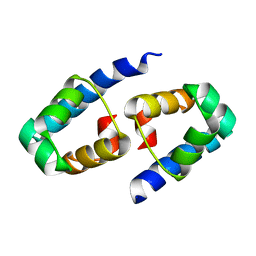 | | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila | | Descriptor: | C.AhdI | | Authors: | McGeehan, J.E, Streeter, S.D, Papapanagiotou, I, Fox, G.C, Kneale, G.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila.
J.Mol.Biol., 346, 2005
|
|
2A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, 40 STRUCTURES | | Descriptor: | C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-06-09 | | Release date: | 1999-01-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
1GNH
 
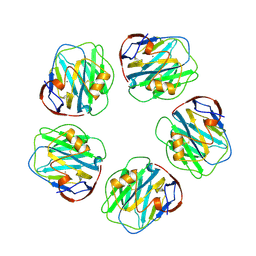 | | HUMAN C-REACTIVE PROTEIN | | Descriptor: | C-REACTIVE PROTEIN, CALCIUM ION | | Authors: | Shrive, A.K, Cheetham, G.M.T, Holden, D, Myles, D.A, Turnell, W.G, Volanakis, J.E, Pepys, M.B, Bloomer, A.C, Greenhough, T.J. | | Deposit date: | 1996-03-01 | | Release date: | 1997-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three dimensional structure of human C-reactive protein.
Nat.Struct.Biol., 3, 1996
|
|
2UUL
 
 | | Crystal structure of C-phycocyanin from Phormidium, Lyngbya spp. (Marine) and Spirulina sp. (Fresh water) shows two different ways of energy transfer between two hexamers. | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN ALPHA CHAIN, C-PHYCOCYANIN BETA CHAIN, ... | | Authors: | Satyanarayana, L, Patel, A, Mishra, S, K Ghosh, P, Suresh, C.G. | | Deposit date: | 2007-03-04 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Phormidium, Lyngbya Spp. (Marine) and Spirulina Sp. (Fresh Water) Shows Two Different Ways of Energy Transfer between Twohexamers.
To be Published
|
|
1GH0
 
 | | CRYSTAL STRUCTURE OF C-PHYCOCYANIN FROM SPIRULINA PLATENSIS | | Descriptor: | C-PHYCOCYANIN ALPHA SUBUNIT, C-PHYCOCYANIN BETA SUBUNIT, PHYCOCYANOBILIN | | Authors: | Liang, D.-C, Chang, W.-R, Wang, X.-Q. | | Deposit date: | 2000-10-29 | | Release date: | 2001-06-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of C-phycocyanin from Spirulina platensis at 2.2 A resolution: a novel monoclinic crystal form for phycobiliproteins in phycobilisomes.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3PVN
 
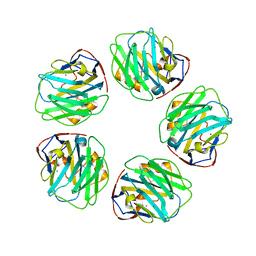 | | Triclinic form of Human C-Reactive Protein in complex with Zinc | | Descriptor: | C-reactive protein, CALCIUM ION, ZINC ION | | Authors: | Guillon, C, Mavoungou Bigouagou, U, Jeannin, P, Delneste, Y, Gouet, P. | | Deposit date: | 2010-12-07 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Staggered Decameric Assembly of Human C-Reactive Protein Stabilized by Zinc Ions Revealed by X-ray Crystallography.
Protein Pept.Lett., 22, 2014
|
|
3PVO
 
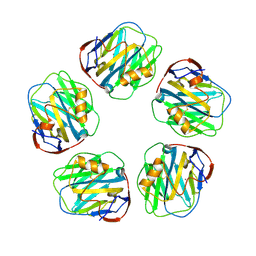 | | Monoclinic form of Human C-Reactive Protein | | Descriptor: | C-Reactive Protein, CALCIUM ION | | Authors: | Guillon, C, Mavoungou Bigouagou, U, Jeannin, P, Delneste, Y, Gouet, P. | | Deposit date: | 2010-12-07 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Staggered Decameric Assembly of Human C-Reactive Protein Stabilized by Zinc Ions Revealed by X-ray Crystallography.
Protein Pept.Lett., 22, 2014
|
|
4RY6
 
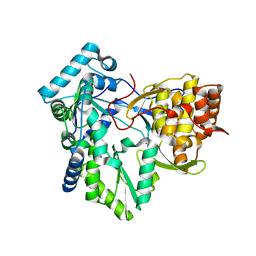 | | C-terminal mutant (W550A) of HCV/J4 RNA polymerase | | Descriptor: | HCV J4 RNA polymerase (NS5B) | | Authors: | Jaeger, J, Cherry, A, Dennis, C. | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
4RY4
 
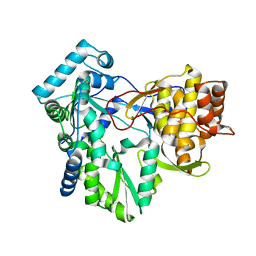 | | C-terminal mutant (Y448F) of HCV/J4 RNA polymerase | | Descriptor: | HCV J4 RNA polymerase (NS5B) | | Authors: | Jaeger, J, Cherry, A, Dennis, C. | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
4RY7
 
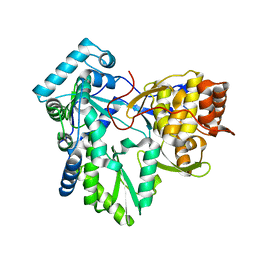 | | C-terminal mutant (D559E) of HCV/J4 RNA polymerase | | Descriptor: | HCV J4 RNA polymerase (NS5B) | | Authors: | Jaeger, J, Cherry, A, Dennis, C. | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
4RY5
 
 | | C-terminal mutant (W550N) of HCV/J4 RNA polymerase | | Descriptor: | HCV J4 RNA polymerase (NS5B), MANGANESE (II) ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Jaeger, J, Cherry, A, Dennis, C. | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Hydrophobic and Charged Residues in the C-Terminal Arm of Hepatitis C Virus RNA-Dependent RNA Polymerase Regulate Initiation and Elongation.
J.Virol., 89, 2015
|
|
1GJI
 
 | | Crystal structure of c-Rel bound to DNA | | Descriptor: | C-REL PROTO-ONCOGENE PROTEIN, IL-2 CD28RE DNA | | Authors: | Huang, D.B, Chen, Y.Q, Ruetsche, M, Phelps, C.B, Ghosh, G. | | Deposit date: | 2001-05-30 | | Release date: | 2002-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2.
Structure, 9, 2001
|
|
