3U06
 
 | | Crystal structure of the kinesin-14 NcdG347D | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, H.-L, Pemble IV, C.W, Endow, S.A. | | Deposit date: | 2011-09-28 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neck-motor interactions trigger rotation of the kinesin stalk.
Sci Rep, 2, 2012
|
|
3U9D
 
 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-24) of drosophila Ciboulot and the C-terminal domain (residues 13-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly.
Embo J., 31, 2012
|
|
3UE5
 
 | | ECP-cleaved Actin in complex with Spir domain D | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chen, C, Phillips, M, Sawaya, M.R, Ralston, C.Y, Quinlan, M.E. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-15 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Multiple Forms of Spire-Actin Complexes and their Functional Consequences.
J.Biol.Chem., 287, 2012
|
|
3CGU
 
 | |
1OFC
 
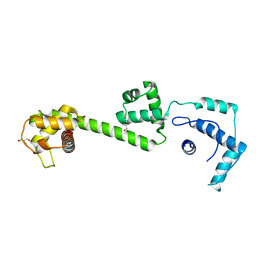 | | nucleosome recognition module of ISWI ATPase | | Descriptor: | 4-deoxy-alpha-D-glucopyranose, GLYCEROL, ISWI PROTEIN, ... | | Authors: | Grune, T, Muller, C.W. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Functional Analysis of a Nucleosome Recognition Module of the Remodeling Factor Iswi
Mol.Cell, 12, 2003
|
|
1OUD
 
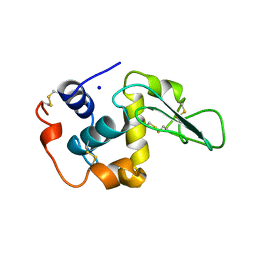 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V121A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1ONQ
 
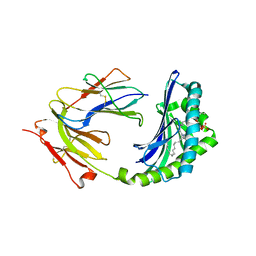 | | Crystal Structure of CD1a in Complex with a Sulfatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Elsliger, M.A, Teyton, L, Wilson, I.A. | | Deposit date: | 2003-02-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A.
Nat.Immunol., 4, 2003
|
|
1OUB
 
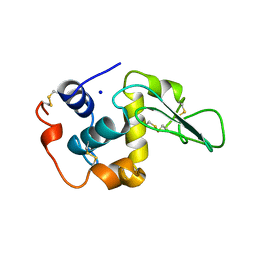 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V100A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUH
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V74A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUF
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V130A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1P9A
 
 | |
1OUG
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V2A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUC
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V110A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUJ
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V99A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUE
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V125A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUI
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V93A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1PZW
 
 | | Crystal structure of the zinc finger associated domain of the Drosophila transcription factor Grauzone | | Descriptor: | Transcription factor grauzone, ZINC ION | | Authors: | Jauch, R, Bourenkov, G.P, Chung, H.-R, Urlaub, H, Reidt, U, Jaeckle, H, Wahl, M.C. | | Deposit date: | 2003-07-14 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein-protein interaction module
STRUCTURE, 11, 2003
|
|
2X6I
 
 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PIK-90 | | Descriptor: | N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
1OGT
 
 | | CRYSTAL STRUCTURE OF HLA-B*2705 COMPLEXED WITH THE VASOACTIVE INTESTINAL PEPTIDE TYPE 1 RECEPTOR (VIPR) PEPTIDE (RESIDUES 400-408) | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Hulsmeyer, M, Fiorillo, M.T, Bettosini, F, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2003-05-13 | | Release date: | 2004-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Dual, HLA-B27 subtype-dependent conformation of a self-peptide.
J. Exp. Med., 199, 2004
|
|
1Q4Q
 
 | | Crystal structure of a DIAP1-Dronc complex | | Descriptor: | Apoptosis 1 inhibitor, Nedd2-like caspase CG8091-PA, ZINC ION | | Authors: | Chai, J, Yan, N, Shi, Y. | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination
Nat.Struct.Biol., 10, 2003
|
|
1NX4
 
 | | The crystal structure of carbapenem synthase (CarC) | | Descriptor: | 2-OXOGLUTARIC ACID, Carbapenem synthase, FE (III) ION | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-08 | | Release date: | 2003-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
2VAS
 
 | | Myosin VI (MD-insert2-CaM, Delta-Insert1) Post-rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
1OC3
 
 | | HUMAN PEROXIREDOXIN 5 | | Descriptor: | BENZOIC ACID, PEROXIREDOXIN 5 | | Authors: | Evrard, C, Declercq, J.-P. | | Deposit date: | 2003-02-05 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Dimeric Oxidized Form of Human Peroxiredoxin 5
J.Mol.Biol., 337, 2004
|
|
1NX8
 
 | | Structure of carbapenem synthase (CarC) complexed with N-acetyl proline | | Descriptor: | 1-ACETYL-L-PROLINE, 2-OXOGLUTARIC ACID, Carbapenem synthase, ... | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
1O70
 
 | |
