7QT6
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z1367324110 | | Descriptor: | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QT5
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z31792168 | | Descriptor: | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8QDY
 
 | |
7QT7
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z4439011520 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(5-tert-butyl-1,2-oxazol-3-yl)-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QGF
 
 | | Cubic Insulin SAD phasing at 14.2 keV | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Nanao, M.H, Basu, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.203 Å) | | Cite: | ID23-2: an automated and high-performance microfocus beamline for macromolecular crystallography at the ESRF.
J.Synchrotron Radiat., 29, 2022
|
|
4P45
 
 | |
8QK1
 
 | |
6BXH
 
 | | Menin in complex with MI-853 | | Descriptor: | 1-{2-[4-(fluoroacetyl)piperazin-1-yl]ethyl}-4-methyl-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Menin in complex with MI-853
To Be Published
|
|
7QGR
 
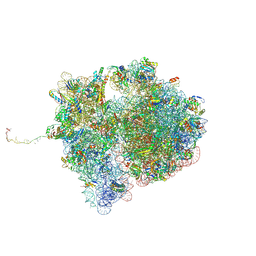 | | Structure of the SmrB-bound E. coli disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
5N0J
 
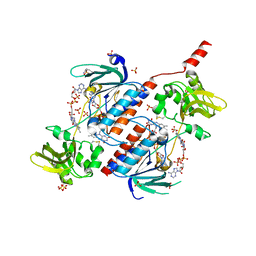 | | Structure of a novel oxidoreductase from Gloeobacter violaceus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Gll2934 protein, SULFATE ION | | Authors: | Buey, R.M, Galindo-Trigo, S, de Pereda, J.M, Balsera, M. | | Deposit date: | 2017-02-03 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Unprecedented pathway of reducing equivalents in a diflavin-linked disulfide oxidoreductase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5C7J
 
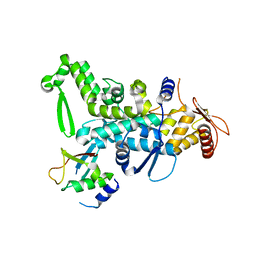 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
6EFH
 
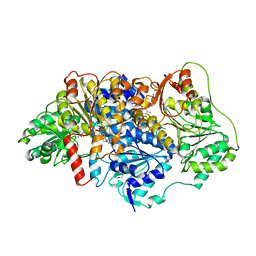 | | Pyruvate decarboxylase from Kluyveromyces lactis soaked with pyruvamide | | Descriptor: | (1S,2S)-1-amino-1,2-dihydroxypropan-1-olate, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Kutter, S, Konig, S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The crystal structures of pyruvate decarboxylase from Kluyveromyces lactis in the absence of ligands and in the presence of the substrate surrogate pyruvamide
To be Published
|
|
4P5Z
 
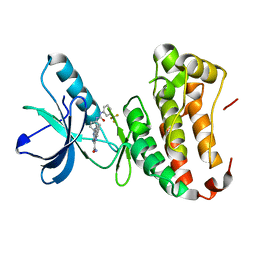 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-[4-({[3-(trifluoromethyl)phenyl]carbamoyl}amino)phenyl]-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-20 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
6N41
 
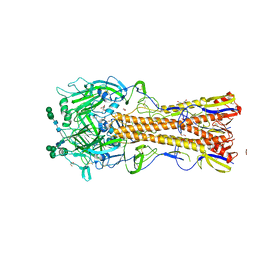 | |
1A7S
 
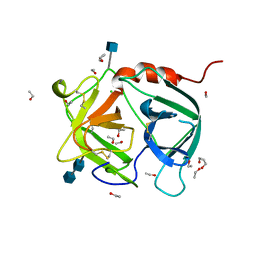 | | ATOMIC RESOLUTION STRUCTURE OF HBP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Karlsen, S, Iversen, L.F, Larsen, I.K, Flodgaard, H.J, Kastrup, J.S. | | Deposit date: | 1998-03-17 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic resolution structure of human HBP/CAP37/azurocidin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
8OOG
 
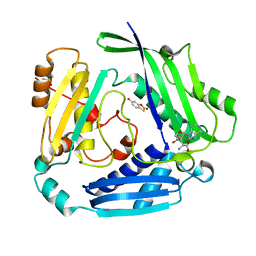 | |
4A9J
 
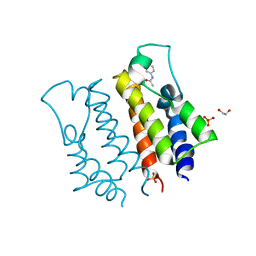 | |
7QVP
 
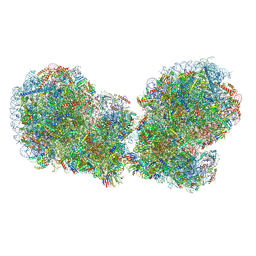 | | Human collided disome (di-ribosome) stalled on XBP1 mRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Denk, T.G, Tesina, P, Beckmann, R. | | Deposit date: | 2022-01-22 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation.
Nat Commun, 13, 2022
|
|
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVL
 
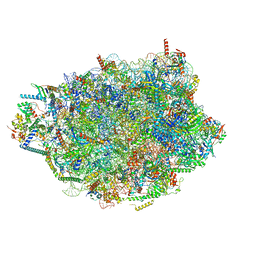 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
7QOW
 
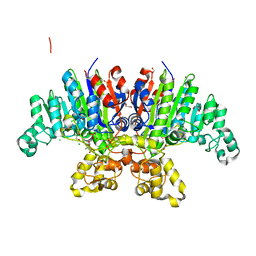 | | Crystal structure of Vibrio alkaline phosphatase in 1.0 M NaCl | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkaline phosphatase, ... | | Authors: | Markusson, S, Hjorleifsson, J.G, Kursula, P, Asgeirsson, B. | | Deposit date: | 2021-12-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Characterization of Functionally Important Chloride Binding Sites in the Marine Vibrio Alkaline Phosphatase.
Biochemistry, 61, 2022
|
|
7QP8
 
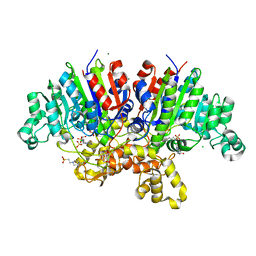 | | Crystal structure of Vibrio alkaline phosphatase with bound HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alkaline phosphatase, CHLORIDE ION, ... | | Authors: | Markusson, S, Hjorleifsson, J.G, Kursula, P, Asgeirsson, B. | | Deposit date: | 2022-01-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Characterization of Functionally Important Chloride Binding Sites in the Marine Vibrio Alkaline Phosphatase.
Biochemistry, 61, 2022
|
|
6GCT
 
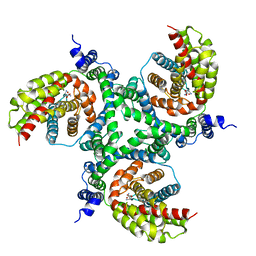 | | cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Garaeva, A.A, Oostergetel, G.T, Gati, C, Guskov, A, Paulino, C, Slotboom, D.J. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of the human neutral amino acid transporter ASCT2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4AGO
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5174 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, TERT-BUTYL [3-(3-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-HYDROXY-5-IODOPHENYL)PROP-2-YN-1-YL]CARBAMATE, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Fersht, A.R, Boeckler, F.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
7BBO
 
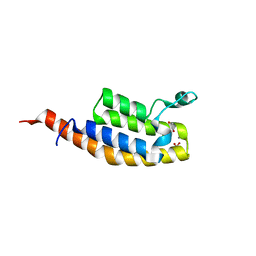 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
