1XGB
 
 | |
1UOK
 
 | | CRYSTAL STRUCTURE OF B. CEREUS OLIGO-1,6-GLUCOSIDASE | | Descriptor: | OLIGO-1,6-GLUCOSIDASE | | Authors: | Watanabe, K, Hata, Y, Kizaki, H, Katsube, Y, Suzuki, Y. | | Deposit date: | 1998-07-28 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 A resolution: structural characterization of proline-substitution sites for protein thermostabilization.
J.Mol.Biol., 269, 1997
|
|
1HAK
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN PLACENTAL ANNEXIN V COMPLEXED WITH K-201 AS A CALCIUM CHANNEL ACTIVITY INHIBITOR | | Descriptor: | 4-[3-{1-(4-BENZYL)PIPERODINYL}PROPIONYL]-7-METHOXY-2,3,4,5-TERTRAHYDRO-1,4-BENZOTHIAZEPINE, ANNEXIN V | | Authors: | Ago, H, Inagaki, E, Miyano, M. | | Deposit date: | 1997-12-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of annexin V with its ligand K-201 as a calcium channel activity inhibitor.
J.Mol.Biol., 274, 1997
|
|
1GGY
 
 | | HUMAN FACTOR XIII WITH YTTERBIUM BOUND IN THE ION SITE | | Descriptor: | PROTEIN (COAGULATION FACTOR XIII), YTTERBIUM (III) ION | | Authors: | Fox, B.A, Yee, V.C, Pederson, L.C, Trong, I.L, Bishop, P.D, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 1998-07-23 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by x-ray crystallography.
J.Biol.Chem., 274, 1999
|
|
1YAC
 
 | |
2A39
 
 | | HUMICOLA INSOLENS ENDOCELLULASE EGI NATIVE STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Sulzenbacher, G, Mackenzie, L, Withers, S.G, Divne, C, Jones, T.A, Woldike, H.F, Schulein, M. | | Deposit date: | 1998-01-30 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335, 1998
|
|
2KZM
 
 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC AND MANGANESE | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*A*CP*GP*C)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE I), ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2ENR
 
 | | CO-CRYSTALS OF DEMETALLIZED CONCANAVALIN A WITH CADMIUM HAVING A CADMIUM ION BOUND IN BOTH THE S1 SITE AND THE S2 SITE | | Descriptor: | CADMIUM ION, CONCANAVALIN A | | Authors: | Bouckaert, J, Loris, R, Wyns, L. | | Deposit date: | 1998-07-14 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Zinc/calcium- and cadmium/cadmium-substituted concanavalin A: interplay of metal binding, pH and molecular packing.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1B8D
 
 | | CRYSTAL STRUCTURE OF A PHYCOUROBILIN-CONTAINING PHYCOERYTHRIN | | Descriptor: | PHYCOERYTHROBILIN, PHYCOUROBILIN, PROTEIN (RHODOPHYTAN PHYCOERYTHRIN (ALPHA CHAIN)), ... | | Authors: | Ritter, S, Hiller, R.G, Wrench, P.M, Welte, W, Diederichs, K. | | Deposit date: | 1999-01-29 | | Release date: | 1999-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phycourobilin-containing phycoerythrin at 1.90-A resolution.
J.Struct.Biol., 126, 1999
|
|
1B72
 
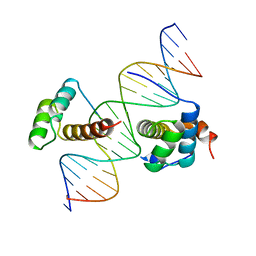 | | PBX1, HOMEOBOX PROTEIN HOX-B1/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*TP*AP*TP*GP*AP*TP*TP*GP*AP*TP*CP*GP*GP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*CP*GP*AP*TP*CP*AP*AP*TP*CP*AP*TP*AP*GP*AP*G)-3'), PROTEIN (HOMEOBOX PROTEIN HOX-B1), ... | | Authors: | Piper, D.E, Batchelor, A.H, Chang, C.-P, Cleary, M.L, Wolberger, C. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation.
Cell(Cambridge,Mass.), 96, 1999
|
|
1XBH
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(L-262) | | Descriptor: | PROTEIN (CYCLO(L-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-02-17 | | Release date: | 1999-02-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
1B9J
 
 | |
1VPP
 
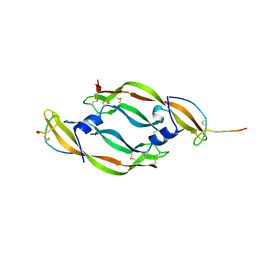 | | COMPLEX BETWEEN VEGF AND A RECEPTOR BLOCKING PEPTIDE | | Descriptor: | PROTEIN (PEPTIDE V108), PROTEIN (VASCULAR ENDOTHELIAL GROWTH FACTOR) | | Authors: | Wiesmann, C, Christinger, H.W, Cochran, A.G, Cunningham, B.C, Fairbrother, W.J, Keenan, C.J, Meng, G, de Vos, A.M. | | Deposit date: | 1998-10-09 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex between VEGF and a receptor-blocking peptide.
Biochemistry, 37, 1998
|
|
1NK2
 
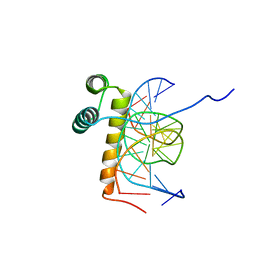 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1BR3
 
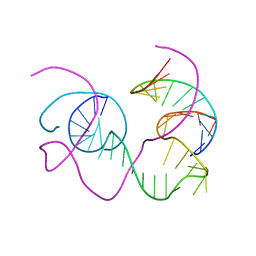 | | CRYSTAL STRUCTURE OF AN 82-NUCLEOTIDE RNA-DNA COMPLEX FORMED BY THE 10-23 DNA ENZYME | | Descriptor: | DNA (10-23 DNA ENZYME), RNA (5'-R(*GP*GP*AP*CP*AP*GP*AP*UP*GP*GP*GP*AP*G)-3') | | Authors: | Nowakowski, J, Shim, P.J, Prasad, G.S, Stout, C.D, Joyce, G.F. | | Deposit date: | 1998-08-13 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10-23 DNA enzyme.
Nat.Struct.Biol., 6, 1999
|
|
2BPY
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(S)-CYCLOPENTYL-1(R)-HYDROXY-3(R)METHYL]-5-[(2(S)-TERTIARY-BUTYLAMINO-CARBONYL)-4-(N1-(2)-(N-METHYLPIPERAZINYL)-3-CHLORO-PYRAZINYL-5-CARBONYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYL-PENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPX
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPZ
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(S)-CYCLOPENTYL-1(R)-HYDROXY-3(R)METHYL]-5-[(2(S)-TERTIARY-BUTYLAMINO-CARBONYL)-4-(N1-(2)-(N-METHYLPIPERAZINYL)-3-CHLORO-PYRAZINYL-5-CARBONYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYL-PENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPW
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | 1-[2-HYDROXY-4-(2-HYDROXY-5-METHYL-CYCLOPENTYLCARBAMOYL)5-PHENYL-PENTYL]-4-(3-PYRIDIN-3-YL-PROPIONYL)-PIPERAZINE-2-CARB OXYLIC ACID TERT-BUTYLAMIDE, HIV-1 PROTEASE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPV
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | 1-[2-HYDROXY-4-(2-HYDROXY-5-METHYL-CYCLOPENTYLCARBAMOYL)5-PHENYL-PENTYL]-4-(3-PYRIDIN-3-YL-PROPIONYL)-PIPERAZINE-2-CARB OXYLIC ACID TERT-BUTYLAMIDE, HIV-1 PROTEASE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3RDN
 
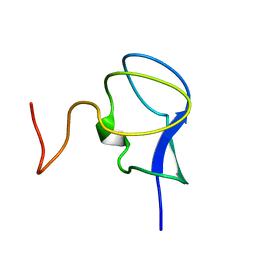 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
1BVY
 
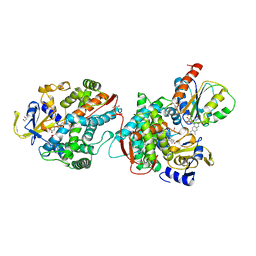 | | COMPLEX OF THE HEME AND FMN-BINDING DOMAINS OF THE CYTOCHROME P450(BM-3) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, PROTEIN (CYTOCHROME P450 BM-3), ... | | Authors: | Sevrioukova, I.F, Li, H, Zhang, H, Peterson, J.A, Poulos, T.L. | | Deposit date: | 1998-09-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1KVP
 
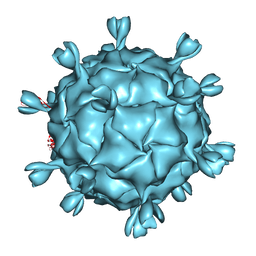 | |
1B33
 
 | | STRUCTURE OF LIGHT HARVESTING COMPLEX OF ALLOPHYCOCYANIN ALPHA AND BETA CHAINS/CORE-LINKER COMPLEX AP*LC7.8 | | Descriptor: | ALLOPHYCOCYANIN, ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Reuter, W, Wiegand, G, Huber, R, Than, M.E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-02-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis at 2.2 A of orthorhombic crystals presents the asymmetry of the allophycocyanin-linker complex, AP.LC7.8, from phycobilisomes of Mastigocladus laminosus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1B9G
 
 | | INSULIN-LIKE-GROWTH-FACTOR-1 | | Descriptor: | PROTEIN (GROWTH FACTOR IGF-1) | | Authors: | De Wolf, E, Gill, R, Geddes, S, Pitts, J, Wollmer, A, Grotzinger, J. | | Deposit date: | 1999-02-11 | | Release date: | 1999-02-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mini IGF-1.
Protein Sci., 5, 1996
|
|
