4K9T
 
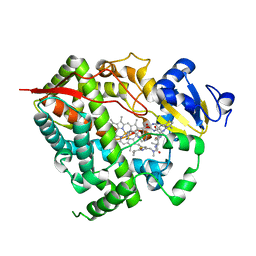 | | Complex of CYP3A4 with a desoxyritonavir analog | | Descriptor: | Cytochrome P450 3A4, N~2~-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-N-(4-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}butyl)-L-valinamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
5L61
 
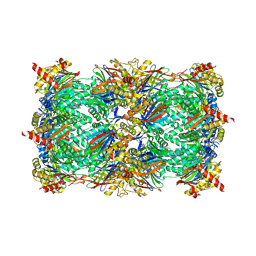 | |
4K9X
 
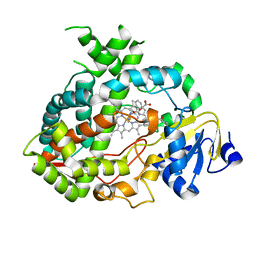 | | Complex of human CYP3A4 with a desoxyritonavir analog | | Descriptor: | 1,3-thiazol-5-ylmethyl [(2R,5R)-5-{[(2S)-2-methylbutanoyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
2C2O
 
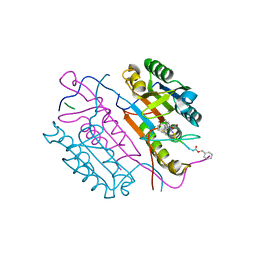 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-14-{4-[BENZYL(METHYL) AMINO]-4-OXOBUTANOYL}-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A, Mikolajczyk, J, Salvesen, G.S, Gruetter, M.G, Powers, J.C. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
4I3Q
 
 | |
2BZ9
 
 | |
4K9V
 
 | | Complex of CYP3A4 with a desoxyritonavir analog | | Descriptor: | 1,3-thiazol-5-ylmethyl [(3S,6S)-6-{[N-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-L-seryl]amino}octan-3-yl]carbamate, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
1SQB
 
 | | Crystal Structure Analysis of Bovine Bc1 with Azoxystrobin | | Descriptor: | Cytochrome b, Cytochrome c1, heme protein, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-18 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex
J.Mol.Biol., 341, 2004
|
|
2B63
 
 | | Complete RNA Polymerase II-RNA inhibitor complex | | Descriptor: | 31-MER, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Kettenberger, H, Eisenfuehr, A, Brueckner, F, Theis, M, Famulok, M, Cramer, P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs
Nat.Struct.Mol.Biol., 13, 2006
|
|
5L8R
 
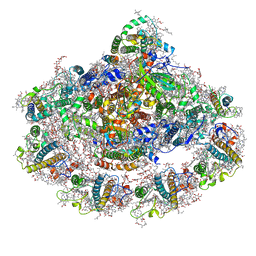 | | The structure of plant photosystem I super-complex at 2.6 angstrom resolution. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Mazor, Y, Borovikova, A, Caspy, I, Nelson, N. | | Deposit date: | 2016-06-08 | | Release date: | 2017-03-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the plant photosystem I supercomplex at 2.6 angstrom resolution.
Nat Plants, 3, 2017
|
|
2GUS
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | Descriptor: | Major outer membrane lipoprotein | | Authors: | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
1SQX
 
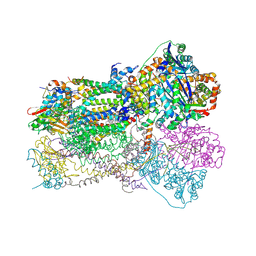 | | Crystal Structure Analysis of Bovine Bc1 with Stigmatellin A | | Descriptor: | Cytochrome b, Cytochrome c1, heme protein, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
4KG8
 
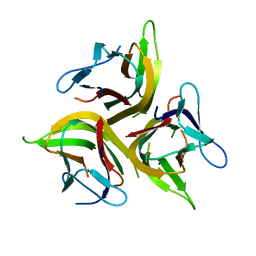 | | Crystal structure of light mutant | | Descriptor: | Tumor necrosis factor ligand superfamily member 14 | | Authors: | Liu, W, Zhan, C, Kumar, P.R, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-28 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4I4H
 
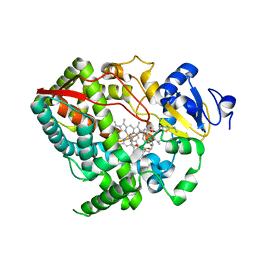 | | Crystal structure of CYP3A4 ligated to pyridine-substituted desoxyritonavir | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, pyridin-3-ylmethyl [(2R,5S)-5-{[N-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-D-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pyridine-Substituted Desoxyritonavir Is a More Potent Inhibitor of Cytochrome P450 3A4 than Ritonavir.
J.Med.Chem., 56, 2013
|
|
4C2M
 
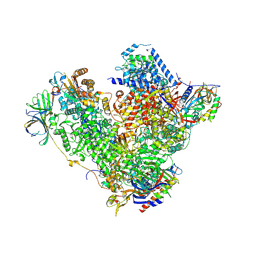 | | Structure of RNA polymerase I at 2.8 A resolution | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Engel, C, Sainsbury, S, Cheung, A.C, Kostrewa, D, Cramer, P. | | Deposit date: | 2013-08-19 | | Release date: | 2013-10-23 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Polymerase I Structure and Transcription Regulation.
Nature, 502, 2013
|
|
2HWF
 
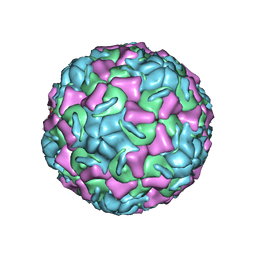 | |
2HWD
 
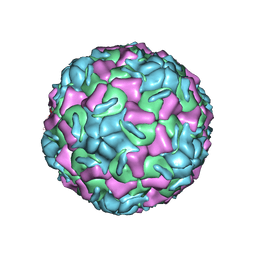 | | A COMPARISON OF THE ANTI-RHINOVIRAL DRUG BINDING POCKET IN HRV14 AND HRV1A | | Descriptor: | 5-(3-(2,6-DICHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PROPYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Kim, K.H, Rossmann, M.G. | | Deposit date: | 1994-01-25 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A.
J.Mol.Biol., 230, 1993
|
|
2HWE
 
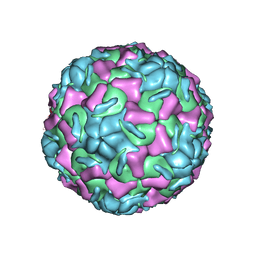 | | A COMPARISON OF THE ANTI-RHINOVIRAL DRUG BINDING POCKET IN HRV14 AND HRV1A | | Descriptor: | 5-(5-(2,6-DICHLORO-4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Kim, K.H, Rossmann, M.G. | | Deposit date: | 1994-01-25 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A.
J.Mol.Biol., 230, 1993
|
|
9BJK
 
 | | Inactive mu opioid receptor bound to Nb6, naloxone and NAM | | Descriptor: | Mu-type opioid receptor, Naloxone, Nalpha-[({(1M)-1-[5-(benzyloxy)pyridin-3-yl]naphthalen-2-yl}sulfanyl)acetyl]-3-methoxy-N,4-dimethyl-L-phenylalaninamide, ... | | Authors: | O'Brien, E.S, Wang, H, Kaavya Krishna, K, Zhang, C, Kobilka, B.K. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | A mu-opioid receptor modulator that works cooperatively with naloxone.
Nature, 631, 2024
|
|
1LOJ
 
 | | Crystal structure of a Methanobacterial Sm-like archaeal protein (SmAP1) bound to uridine-5'-monophosphate (UMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, URIDINE, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Mura, C, Kozhukhovsky, A, Eisenberg, D. | | Deposit date: | 2002-05-06 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs)
Protein Sci., 12, 2003
|
|
5EGL
 
 | | The structural and biochemical characterization of acyl-coa hydrolase from Staphylococcus aureus in complex with Butyryl Coenzyme A, Coenzyme A, and Coenzyme A disulfide | | Descriptor: | Acyl CoA Hydrolase, Butyryl Coenzyme A, COENZYME A, ... | | Authors: | Khandokar, Y.B, Srivastava, P.S, Forwood, J.K. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural and biochemical characterization of acyl-coa hydrolase from Staphylococcus aureus
To Be Published
|
|
5ENJ
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-14 N11530 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | MAGNESIUM ION, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-~{N}-methyl-ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
8CY5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-{3-[1-(tert-butoxycarbonyl)piperidin-4-yl]propyl}adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXX
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 6 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY3
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-[3-(4-aminophenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
