4QXU
 
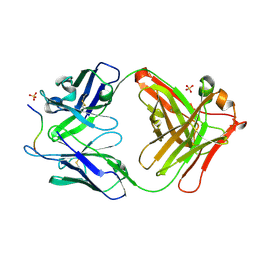 | | Novel Inhibition Mechanism of Membrane Metalloprotease by an Exosite-Swiveling Conformational antibody | | Descriptor: | Matrix metalloproteinase-14, SULFATE ION, anti_MT1-MMP Heavy chain, ... | | Authors: | Udi, Y, Grossman, M, Solomonov, I, Dym, O, Rozenberg, H, Moreno, v, Cuiniasse, P, Dive, V, Arroyo, A.G, Sagi, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2014-07-22 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
4QY8
 
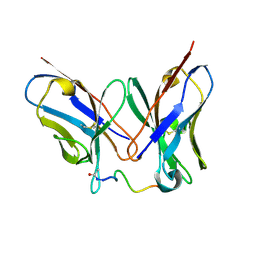 | | Crystal Structure of anti-MSP2 Fv fragment (mAb6D8) in complex with 3D7-MSP2 14-30 | | Descriptor: | Fv fragment(mAb6D8) heavy chain, Fv fragment(mAb6D8) light chain, Merozoite surface antigen 2 | | Authors: | Morales, R.A.V, MacRaild, C.A, Seow, J, Bankala, K, Drinkwater, N, McGowan, S, Rouet, R, Christ, D, Anders, R.F, Norton, R.S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.353 Å) | | Cite: | Structural basis for epitope masking and strain specificity of a conserved epitope in an intrinsically disordered malaria vaccine candidate.
Sci Rep, 5, 2015
|
|
6VXV
 
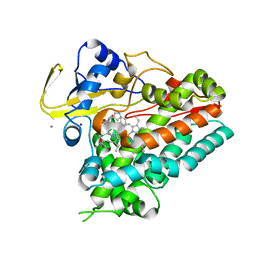 | | Crystal structure of cyclo-L-Trp-L-Pro-bound cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VW2
 
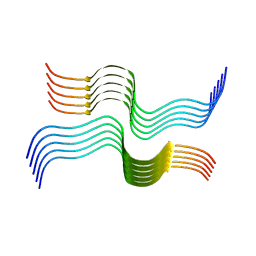 | | Cryo-EM structure of human islet amyloid polypeptide (hIAPP, or amylin) fibrils | | Descriptor: | Islet amyloid polypeptide (SUMO-tagged) | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2020-02-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure and inhibitor design of human IAPP (amylin) fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4QYO
 
 | | Crystal Structure of anti-MSP2 Fv fragment (mAb6D8)in complex with MSP2 14-22 | | Descriptor: | Fv fragment(mAb6D8) heavy chain, Fv fragment(mAb6D8) light chain, Merozoite surface antigen 2 | | Authors: | Morales, R.A.V, MacRaild, C.A, Seow, J, Bankala, K, Drinkwater, N, McGowan, S, Rouet, R, Christ, D, Anders, R.F, Norton, R.S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.208 Å) | | Cite: | Structural basis for epitope masking and strain specificity of a conserved epitope in an intrinsically disordered malaria vaccine candidate.
Sci Rep, 5, 2015
|
|
6ZJN
 
 | | Respiratory complex I from Thermus thermophilus, NADH dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
8FRN
 
 | | Acinetobacter baylyi LptB2FG bound to lipopolysaccharide and Zosurabalpin | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, DODECYL-BETA-D-MALTOSIDE, LPS export ABC transporter permease LptG, ... | | Authors: | Pahil, K.S, Gilman, M.S.A, Kruse, A.C, Kahne, D. | | Deposit date: | 2023-01-07 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A new antibiotic traps lipopolysaccharide in its intermembrane transporter.
Nature, 625, 2024
|
|
4RQL
 
 | |
4RRT
 
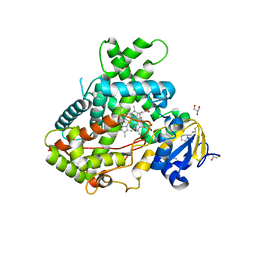 | |
6ZJY
 
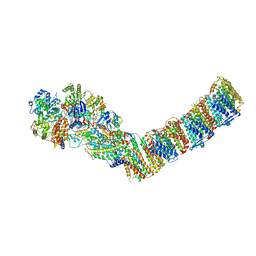 | | Respiratory complex I from Thermus thermophilus, NAD+ dataset, minor state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH-quinone oxidoreductase subunit 1, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6LQR
 
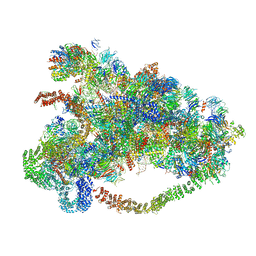 | |
4PDW
 
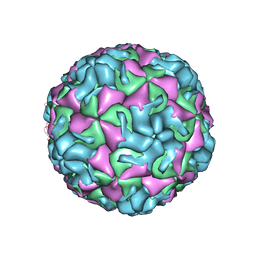 | | A benzonitrile analogue inhibits rhinovirus replication | | Descriptor: | 4-[(4,5-dimethoxy-2-nitrophenyl)acetyl]benzonitrile, Capsid protein VP4/VP2, GLYCEROL, ... | | Authors: | Querol-Audi, J, Guerra, P, Lacroix, C, Roche, M, Franco, D, Froeyen, M, Terme, T, Vanelle, P, Neyts, J, Leyssen, P, Verdaguer, N. | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel benzonitrile analogue inhibits rhinovirus replication.
J.Antimicrob.Chemother., 69, 2014
|
|
6B8H
 
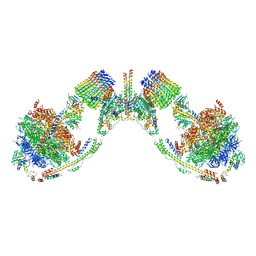 | | Mosaic model of yeast mitochondrial ATP synthase monomer | | Descriptor: | AATP synthase subunit g, ATP synthase catalytic sector F1 epsilon subunit, ATP synthase protein 8, ... | | Authors: | Guo, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2017-10-07 | | Release date: | 2018-01-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic model for the dimeric FO region of mitochondrial ATP synthase.
Science, 358, 2017
|
|
6LQV
 
 | |
6LQT
 
 | |
6LQQ
 
 | |
4NY4
 
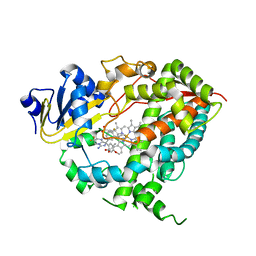 | | Crystal structure of CYP3A4 in complex with an inhibitor | | Descriptor: | (8R)-3,3-difluoro-8-[4-fluoro-3-(pyridin-3-yl)phenyl]-8-(4-methoxy-3-methylphenyl)-2,3,4,8-tetrahydroimidazo[1,5-a]pyrimidin-6-amine, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Branden, G, Sjogren, T, Xue, Y. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-based ligand design to overcome CYP inhibition in drug discovery projects.
Drug Discov Today, 19, 2014
|
|
6S7M
 
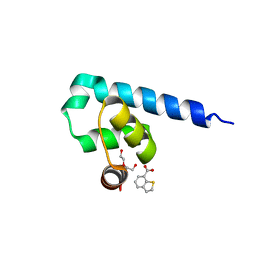 | |
4PKO
 
 | | Crystal structure of the Football-shaped GroEL-GroES2-(ADPBeFx)14 complex | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fei, X, Ye, X, Laronde-Leblanc, N, Lorimer, G.H. | | Deposit date: | 2014-05-15 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
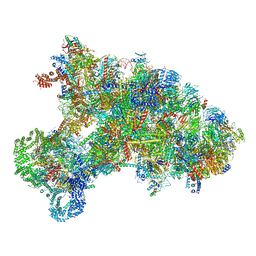
6KE6
 
 | |
3B63
 
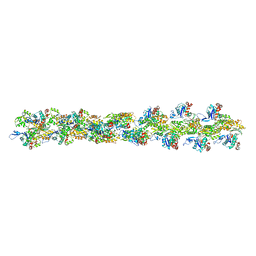 | | Actin filament model in the extended form of acromsomal bundle in the Limulus sperm | | Descriptor: | Actin | | Authors: | Cong, Y, Topf, M, Sali, A, Matsudaira, P, Dougherty, M, Chiu, W, Schmid, M.F. | | Deposit date: | 2007-10-26 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Crystallographic conformers of actin in a biologically active bundle of filaments.
J.Mol.Biol., 375, 2008
|
|
2YU9
 
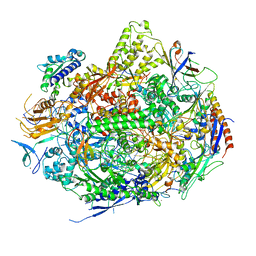 | | RNA polymerase II elongation complex in 150 mm MG+2 with UTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2007-04-06 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Transcription: Role of the Trigger Loop in Substrate Specificity and Catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
6C0F
 
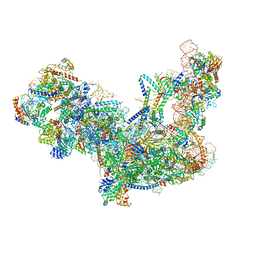 | | Yeast nucleolar pre-60S ribosomal subunit (state 2) | | Descriptor: | 5.8S rRNA, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2017-12-29 | | Release date: | 2018-03-14 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
2AYN
 
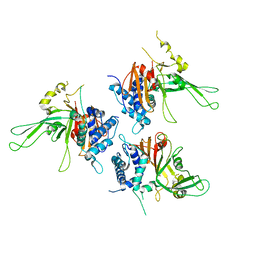 | | Structure of USP14, a proteasome-associated deubiquitinating enzyme | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Hu, M, Li, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-09-07 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14.
Embo J., 24, 2005
|
|
3BJ9
 
 | | Crystal structure of the Surrogate Light Chain Variable Domain VpreBJ | | Descriptor: | 1,2-ETHANEDIOL, Immunoglobulin iota chain, Immunoglobulin lambda-like polypeptide 1 | | Authors: | Morstadt, L.M, Bohm, A.A, Stollar, B.D, Baleja, J.D. | | Deposit date: | 2007-12-03 | | Release date: | 2008-03-04 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering and characterization of a single chain surrogate light chain variable domain.
Protein Sci., 17, 2008
|
|
