9G09
 
 | |
1M3S
 
 | | Crystal structure of YckF from Bacillus subtilis | | Descriptor: | Hypothetical protein yckf | | Authors: | Sanishvili, R, Wu, R, Kim, D.E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-06-28 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Bacillus subtilis YckF: structural and functional evolution.
J.Struct.Biol., 148, 2004
|
|
1MQ6
 
 | | Crystal Structure of 3-chloro-N-[4-chloro-2-[[(5-chloro-2-pyridinyl)amino]carbonyl]-6-methoxyphenyl]-4-[[(4,5-dihydro-2-oxazolyl)methylamino]methyl]-2-thiophenecarboxamide Complexed with Human Factor Xa | | Descriptor: | 3-CHLORO-N-[4-CHLORO-2-[[(5-CHLORO-2-PYRIDINYL)AMINO]CARBONYL]-6-METHOXYPHENYL]-4-[[(4,5-DIHYDRO-2-OXAZOLYL)METHYLAMINO]METHYL]-2-THIOPHENECARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X HEAVY CHAIN, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2002-09-13 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Two Potent Nonamidine Inhibitors Bound to Factor Xa
Biochemistry, 41, 2002
|
|
9G79
 
 | |
7R62
 
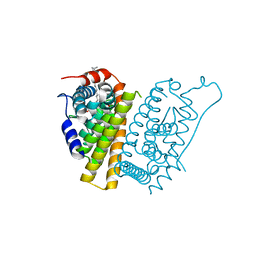 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with a Desmethyl ICI164,384 Derivative | | Descriptor: | 11-[3,17beta-dihydroxyestra-1,3,5(10)-trien-7beta-yl]-N-methyl-N-propylundecanamide, Estrogen receptor | | Authors: | Diennet, M, El Ezzy, M, Thiombane, K, Cotnoir-White, D, Poupart, J, Gao, Z, Mendoza Sanchez, R, Marinier, A, Gleason, J, Greene, G.L, Mader, S.C, Fanning, S.W. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain in Complex with a Desmethyl ICI164,384 Derivative
To Be Published
|
|
1M4H
 
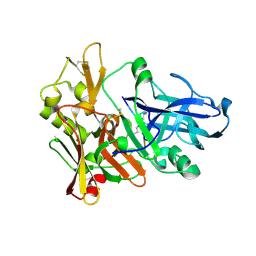 | | Crystal Structure of Beta-secretase complexed with Inhibitor OM00-3 | | Descriptor: | Inhibitor OM00-3, beta-Secretase | | Authors: | Hong, L, Turner, R.T, Koelsch, G, Ghosh, A.K, Tang, J. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Memapsin 2 (beta-Secretase) in Complex with Inhibitor OM00-3
Biochemistry, 41, 2002
|
|
1M55
 
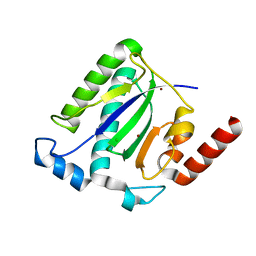 | | Catalytic domain of the Adeno Associated Virus type 5 Rep protein | | Descriptor: | CHLORIDE ION, Rep protein, ZINC ION | | Authors: | Hickman, A.B, Ronning, D.R, Kotin, R.M, Dyda, F. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep.
Mol.Cell, 10, 2002
|
|
1MKP
 
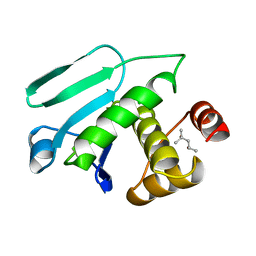 | | CRYSTAL STRUCTURE OF PYST1 (MKP3) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PYST1 | | Authors: | Stewart, A.E, Dowd, S, Keyse, S, Mcdonald, N.Q. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the MAPK phosphatase Pyst1 catalytic domain and implications for regulated activation.
Nat.Struct.Biol., 6, 1999
|
|
9GA2
 
 | | MtUvrA2 dimer empty | | Descriptor: | UvrABC system protein A, ZINC ION | | Authors: | Genta, M, Capelli, R, Ferrara, G, Rizzi, M, Rossi, F, Jeruzalmi, D, Bolognesi, M, Chaves-Sanjuan, A, Miggiano, R. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Mechanistic understanding of UvrA damage detection and lesion hand-off to UvrB in Nucleotide Excision Repair.
Nat Commun, 16, 2025
|
|
1MJL
 
 | |
1M5C
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
7RP9
 
 | | X-ray crystal structure of OXA-24/40 V130D in complex with imipenem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, BICARBONATE ION, Beta-lactamase, ... | | Authors: | Powers, R.A, Mitchell, J.M, June, C.M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Conformational flexibility in carbapenem hydrolysis drives substrate specificity of the class D carbapenemase OXA-24/40.
J.Biol.Chem., 298, 2022
|
|
9G5D
 
 | | Assembly intermediate of human mitochondrial ribosome small subunit (State C) | | Descriptor: | 12S mitochondrial rRNA, 12S rRNA N4-methylcytidine (m4C) methyltransferase, 28S ribosomal protein S10, ... | | Authors: | Finke, A.F, Heinrichs, M, Aibara, S, Richter-Dennerlein, R, Hillen, H.S. | | Deposit date: | 2024-07-16 | | Release date: | 2025-04-23 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Coupling of ribosome biogenesis and translation initiation in human mitochondria.
Nat Commun, 16, 2025
|
|
9G6J
 
 | | The structure of the Candida albicans ribosome with tRNA-fMet, mRNA, and compounds (GEN and MFQ) with strong density for the P-site tRNA | | Descriptor: | (11R,12S)- Mefloquine, 18S rRNA, 25S rRNA, ... | | Authors: | Kolosova, O, Zgadzay, Y, Jenner, L.B, Guskov, A, Yusupov, M. | | Deposit date: | 2024-07-18 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Mechanism of read-through enhancement by aminoglycosides and mefloquine.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1M5L
 
 | |
9GA4
 
 | | MtUvrA2UvrB2 bound to damaged oligonucleotide | | Descriptor: | DNA, UvrABC system protein A, UvrABC system protein B, ... | | Authors: | Genta, M, Capelli, R, Ferrara, G, Rizzi, M, Rossi, F, Jeruzalmi, D, Bolognesi, M, Chaves-Sanjuan, A, Miggiano, R. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic understanding of UvrA damage detection and lesion hand-off to UvrB in Nucleotide Excision Repair.
Nat Commun, 16, 2025
|
|
1M65
 
 | | YCDX PROTEIN | | Descriptor: | Hypothetical protein ycdX, SODIUM ION, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-07-12 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of the Escherichia coli YcdX protein reveals a trinuclear zinc active site
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
7R57
 
 | |
9GOS
 
 | | CryoEM structure of the native Chlamydomonas reinhardtii Flagella Membrane Glycoprotein 1B. | | Descriptor: | Flagella Membrane Glycoprotein 1B | | Authors: | Nievergelt, A.P, Hoepfner, L.M, Matrino, F, Scholz, M, Foster, H.E, Rodenfels, J, von Appen, A, Hippler, M, Pigino, G. | | Deposit date: | 2024-09-06 | | Release date: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CryoEM structure of the native Chlamydomonas reinhardtii Flagella Membrane Glycoprotein 1B.
To Be Published
|
|
1LUX
 
 | |
1LVA
 
 | |
9GK1
 
 | |
7R8P
 
 | |
1LVJ
 
 | |
9GLH
 
 | | Crystal Structure of UFC1 T106S | | Descriptor: | SULFATE ION, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Kumar, M, Banerjee, S, Wiener, R. | | Deposit date: | 2024-08-27 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.115 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
