9G7H
 
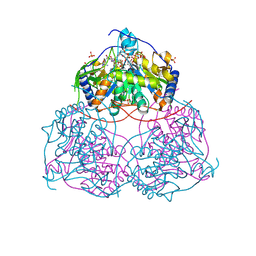 | | Human Sirt6 in complex with ADP-ribose and the inhibitor 2-Pr | | Descriptor: | 2,4-bis(oxidanylidene)-1-[2-oxidanylidene-2-[[(2S)-3-oxidanylidene-3-(propylamino)-2-[3-(3-pyridin-3-yl-1,2,4-oxadiazol-5-yl)propanoylamino]propyl]amino]ethyl]pyrimidine-5-carboxamide, NAD-dependent protein deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2024-07-21 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidating the Unconventional Binding Mode of a DNA-Encoded Library Hit Provides a Blueprint for Sirtuin 6 Inhibitor Development.
Chemmedchem, 2024
|
|
5AM7
 
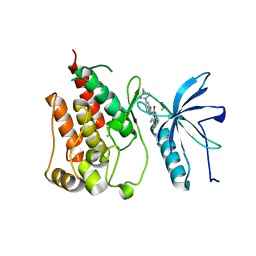 | | FGFR1 mutant with an inhibitor | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, CHLORIDE ION, FIBROBLAST GROWTH FACTOR RECEPTOR 1 | | Authors: | Bunney, T.D, Wan, S, Thiyagarajan, N, Sutto, L, Williams, S.V, Ashford, P, Koss, H, Knowles, M.A, Gervasio, F.L, Coveney, P.V, Katan, M. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study
Ebiomedicine, 2, 2015
|
|
5AM6
 
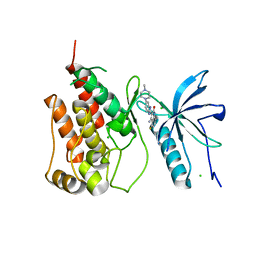 | | Native FGFR1 with an inhibitor | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, CHLORIDE ION, FIBROBLAST GROWTH FACTOR RECEPTOR 1 | | Authors: | Bunney, T.D, Wan, S, Thiyagarajan, N, Sutto, L, Williams, S.V, Ashford, P, Koss, H, Knowles, M.A, Gervasio, F.L, Coveney, P.V, Katan, M. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study
Ebiomedicine, 2, 2015
|
|
6G0V
 
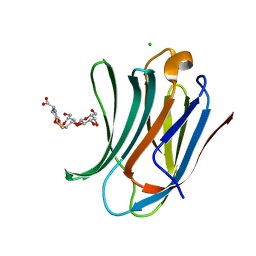 | | Human Galectin-3 in complex with a TF tumor-associated antigen mimetic | | Descriptor: | (3~{R},5~{R},6~{S},7~{S},8~{R},13~{S})-5-(hydroxymethyl)-7-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-oxidanyl-11-oxidanylidene-2,4-dioxa-9-thia-12-azatricyclo[8.4.0.0^{3,8}]tetradec-1(10)-ene-13-carboxylic acid, CHLORIDE ION, Galectin-3 | | Authors: | Trovao, F.G, Santarsia, S, Carvalho, A.L. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular Recognition of a Thomsen-Friedenreich Antigen Mimetic Targeting Human Galectin-3.
ChemMedChem, 13, 2018
|
|
1UW4
 
 | |
8BGD
 
 | | Structure of Mpro from SARS-CoV-2 in complex with FGA147 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-4-methyl-1-[[(2~{S})-4-nitro-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | Peptidyl Nitroalkenes Inhibit SARS-CoV-2 Main Protease and Block Virus Replication
To Be Published
|
|
8BFQ
 
 | |
3IL1
 
 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, IDRA-21 | | Descriptor: | (3S)-7-chloro-3-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3ILT
 
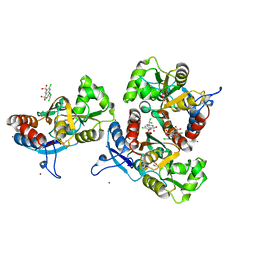 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, trichlormethiazide | | Descriptor: | 6-CHLORO-3-(DICHLOROMETHYL)-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE-7-SULFONAMIDE 1,1-DIOXIDE, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
1X1Y
 
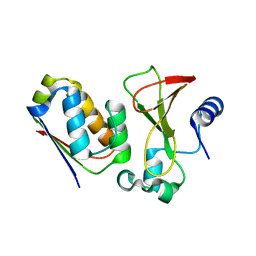 | |
1X1U
 
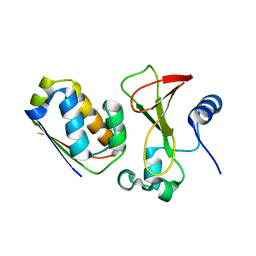 | |
1X1X
 
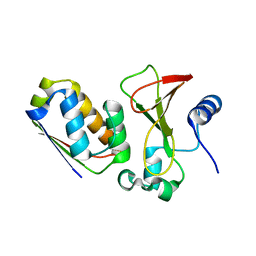 | |
3ILU
 
 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydroflumethiazide | | Descriptor: | 6-(trifluoromethyl)-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
7PZ1
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH8535 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-bromanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(3-methoxy-4-methyl-phenyl)piperidine-1-carboxamide, GLYCEROL, ... | | Authors: | Scaletti, E.R, Helleday, T, Stenmark, P. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
8BXP
 
 | |
1X1W
 
 | |
8C1X
 
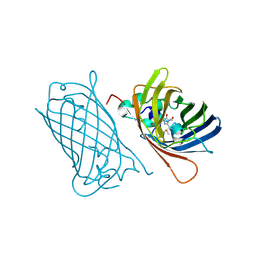 | |
6B3E
 
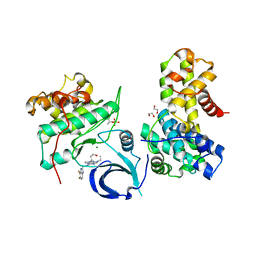 | | Crystal structure of human CDK12/CyclinK in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S)-1-(6-{[(4,5-difluoro-1H-benzimidazol-2-yl)methyl]amino}-9-ethyl-9H-purin-2-yl)piperidin-2-yl]ethan-1-ol, Cyclin-K, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-09-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure-Based Design of Selective Noncovalent CDK12 Inhibitors.
ChemMedChem, 13, 2018
|
|
5FP0
 
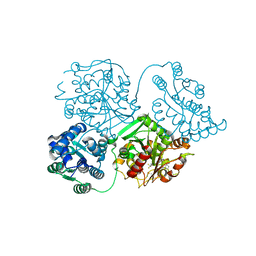 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, N-cyclopentyl-2-[4-(trifluoromethyl)phenyl]-3H-benzimidazole-4-sulfonamide | | Authors: | Xue, Y, Olsson, T, Johansson, C.A, Oster, L, Beisel, H.G, Rohman, M, Karis, D, Backstrom, S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment Screening of Soluble Epoxide Hydrolase for Lead Generation-Structure-Based Hit Evaluation and Chemistry Exploration.
Chemmedchem, 11, 2016
|
|
7O7B
 
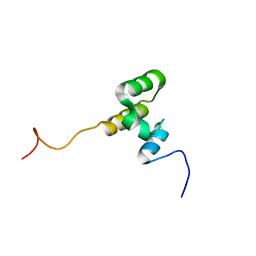 | |
6UUN
 
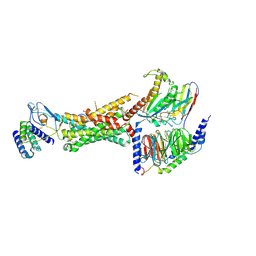 | | CryoEM Structure of the active Adrenomedullin 1 receptor G protein complex with adrenomedullin peptide | | Descriptor: | ADM, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-10-30 | | Release date: | 2020-03-25 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
4N6R
 
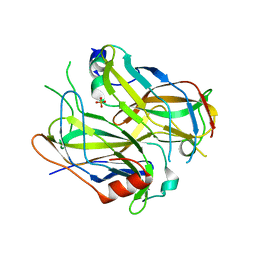 | | Crystal structure of VosA-VelB-complex | | Descriptor: | SULFATE ION, VelB, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
6QHJ
 
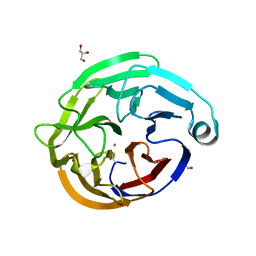 | | High-resolution crystal structure of calcium- and sodium-bound mouse Olfactomedin-1 beta-propeller domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Pronker, M.F, van den Hoek, H.G, Janssen, B.J.C. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol, 20, 2019
|
|
4CEM
 
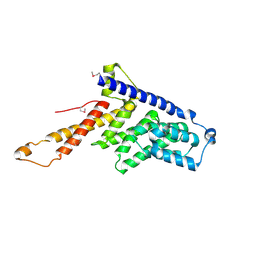 | | Crystal structure of the first MIF4G domain of human nonsense mediated decay factor UPF2 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 2 | | Authors: | Clerici, M, Deniaud, A, Boehm, V, Gehring, N.H, Schaffitzel, C, Cusack, S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-11 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Analysis of the Three Mif4G Domains of Nonsense-Mediated Decay Factor Upf2.
Nucleic Acids Res., 42, 2014
|
|
4CEK
 
 | | Crystal structure of the second MIF4G domain of human nonsense mediated decay factor UPF2 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 2 | | Authors: | Clerici, M, Deniaud, A, Boehm, V, Gehring, N.H, Schaffitzel, C, Cusack, S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Functional Analysis of the Three Mif4G Domains of Nonsense-Mediated Decay Factor Upf2.
Nucleic Acids Res., 42, 2014
|
|
