4NNY
 
 | | Crystal structure of non-phosphorylated form of PKD2 phosphopeptide bound to HLA-A2 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, GLYCEROL, ... | | Authors: | Mohammed, F, Stones, D.H, Willcox, B.E. | | Deposit date: | 2013-11-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status.
Oncotarget, 8, 2017
|
|
4NO5
 
 | | Crystal structure of non-phosphorylated form of AMPD2 phosphopeptide bound to HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, AMP deaminase 2, Beta-2-microglobulin, ... | | Authors: | Mohammed, F, Stones, D.H, Willcox, B.E. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status.
Oncotarget, 8, 2017
|
|
4NYW
 
 | | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand | | Descriptor: | (3R)-N-[3-(3,4-dihydroquinolin-1(2H)-yl)propyl]-3-methyl-2-oxo-1,2,3,4-tetrahydroquinoxaline-5-carboxamide, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Rooney, T.P.C, Fedorov, O, Martin, S, Monteiro, O.P, Conway, S.J, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand
TO BE PUBLISHED
|
|
4NOX
 
 | | Structure of the nine-bladed beta-propeller of eIF3b | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit B | | Authors: | Liu, Y, Neumann, P, Kuhle, B, Monecke, T, Ficner, R. | | Deposit date: | 2013-11-20 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.722 Å) | | Cite: | Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit
Structure, 22, 2014
|
|
4EH4
 
 | | Human p38 MAP kinase in complex with NP-F3 and RL87 | | Descriptor: | Mitogen-activated protein kinase 14, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine, phenyl(piperidin-1-yl)methanone | | Authors: | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | Deposit date: | 2012-04-02 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
4EJF
 
 | |
4NTO
 
 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C2 ceramide-1-phosphate (d18:1/2:0) at 2.15 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NU5
 
 | | Crystal Structure of PTDH R301A | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase | | Authors: | Nair, S.K, Chekan, J.R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4EM3
 
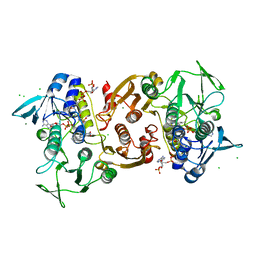 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor MeVS-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wallace, B.D, Edwards, J.S, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4NVN
 
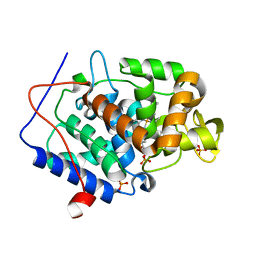 | | Predicting protein conformational response in prospective ligand discovery | | Descriptor: | 2,3-dihydrobenzo[h][1,6]naphthyridin-4(1H)-one, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery.
Nat Chem, 6, 2014
|
|
4NCK
 
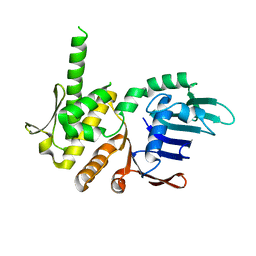 | | Crystal Structure of Pyrococcus furiosis Rad50 R797G mutation | | Descriptor: | CHLORIDE ION, DNA double-strand break repair Rad50 ATPase, MAGNESIUM ION, ... | | Authors: | Classen, S, Williams, G.J, Arvai, A.S, Williams, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling.
Embo J., 33, 2014
|
|
4EQ1
 
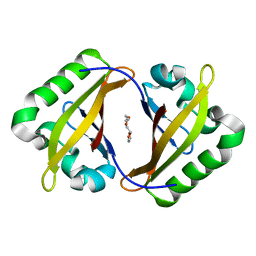 | | Crystal Structure of the ARNT PAS-B homodimer | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Aryl hydrocarbon receptor nuclear translocator | | Authors: | Gardner, K.H, Key, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Regulating the ARNT/TACC3 Axis: Multiple Approaches to Manipulating Protein/Protein Interactions with Small Molecules.
Acs Chem.Biol., 8, 2013
|
|
4ER7
 
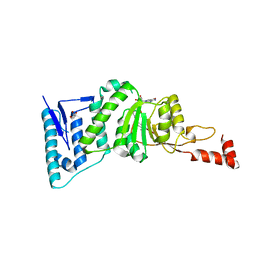 | | Crystal Structure of human DOT1L in complex with inhibitor SGC0947 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4ES1
 
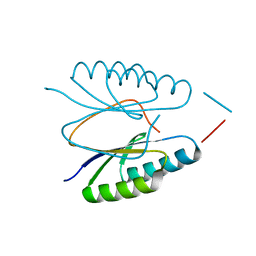 | |
4NF1
 
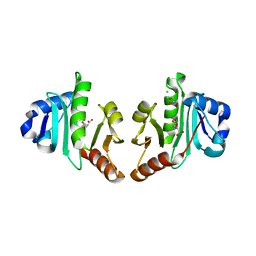 | | Structure of N-acetyltransferase domain of X. fastidiosa NAGS/K without his-tag | | Descriptor: | Acetylglutamate kinase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2013-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3986 Å) | | Cite: | Structures of the N-acetyltransferase domain of Xylella fastidiosaN-acetyl-L-glutamate synthase/kinase with and without a His tag bound to N-acetyl-L-glutamate.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4NGQ
 
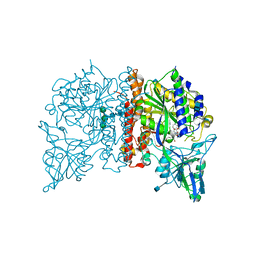 | | Crystal Structure of Glutamate Carboxypeptidase II in a complex with urea-based inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Pachl, P. | | Deposit date: | 2013-11-02 | | Release date: | 2014-06-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Rational design of urea-based glutamate carboxypeptidase II (GCPII) inhibitors as versatile tools for specific drug targeting and delivery.
Bioorg.Med.Chem., 22, 2014
|
|
4EXT
 
 | |
4NIM
 
 | |
4NJ4
 
 | | Crystal Structure of Human ALKBH5 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation.
Nucleic Acids Res., 42, 2014
|
|
4ER8
 
 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
4ERQ
 
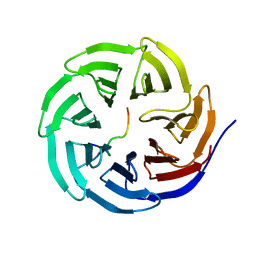 | | X-ray structure of WDR5-MLL2 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL2, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4NP3
 
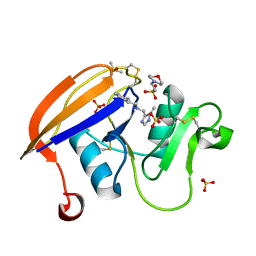 | | Crystal structure of the murine cd44 hyaluronan binding domain complex with a small molecule | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(4-methyl-1H-imidazol-5-yl)methyl]-1,2,3,4-tetrahydroisoquinolin-8-amine, CD44 antigen, ... | | Authors: | Liu, L.K, Finzel, B. | | Deposit date: | 2013-11-20 | | Release date: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Identification of an Inducible Binding Site on Cell Surface Receptor CD44 for the Design of Protein-Carbohydrate Interaction Inhibitors.
J.Med.Chem., 57, 2014
|
|
4EWG
 
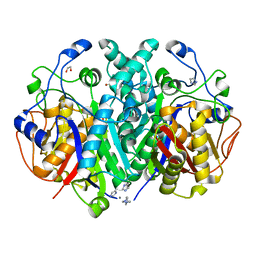 | |
4NPS
 
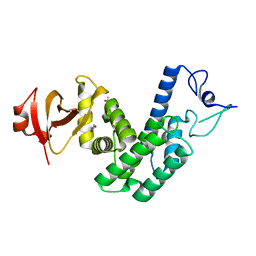 | | Crystal Structure of Bep1 protein (VirB-translocated Bartonella effector protein) from Bartonella clarridgeiae | | Descriptor: | ACETATE ION, Bartonella effector protein (Bep) substrate of VirB T4SS | | Authors: | Dranow, D.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolutionary Diversification of Host-Targeted Bartonella Effectors Proteins Derived from a Conserved FicTA Toxin-Antitoxin Module.
Microorganisms, 9, 2021
|
|
4NUF
 
 | | Crystal Structure of SHP/EID1 | | Descriptor: | EID1 peptide, Maltose ABC transporter periplasmic protein, Nuclear receptor subfamily 0 group B member 2 chimeric construct, ... | | Authors: | Zhi, X, Zhou, X.E, He, Y, Zechner, C, Suino-Powell, K.M, Kliewer, S.A, Melcher, K, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2013-12-03 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into gene repression by the orphan nuclear receptor SHP.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
