5RZI
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z48852953 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-N-{[(2R)-oxolan-2-yl]methyl}-1,3-thiazol-2-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
7GT9
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMSOA000463b | | Descriptor: | (3R)-4-oxo-3,4-dihydro-2H-1-benzopyran-3-carbonitrile, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
4DX5
 
 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, Acriflavine resistance protein B, DARPIN, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4H3Z
 
 | |
7JVP
 
 | | Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-3-methyl-1-(3-methylphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JV5
 
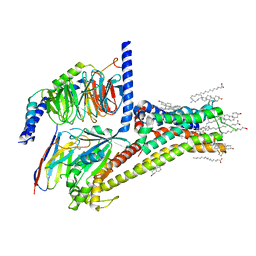 | | Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
5UNG
 
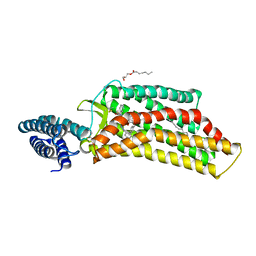 | | XFEL structure of human angiotensin II type 2 receptor (Orthorhombic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl] methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide, ... | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
5E9C
 
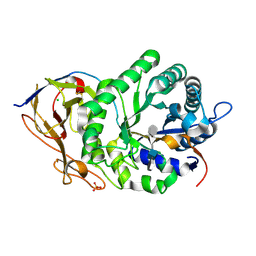 | | Crystal structure of human heparanase in complex with heparin tetrasaccharide dp4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5RHR
 
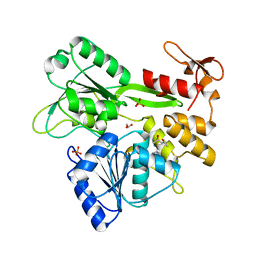 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z1348559502 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-(methylamino)cyclopentane-1-carboxamide, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RYM
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z31217395 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
4E1A
 
 | | Phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis at 1.62A resolution | | Descriptor: | GLYCEROL, Phosphopantetheine adenylyltransferase | | Authors: | Timofeev, V.I, Smirnova, E.A, Chupova, L.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2012-03-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray study of the conformational changes in the molecule of phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis during the catalyzed reaction.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4CF1
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2s)-6-[[(1r,2s)-2-(5-azanylpentanoylamino)-2,3-dihydro-1h-inden-1-yl]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-13 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
1IQI
 
 | | Human coagulation factor Xa in complex with M55125 | | Descriptor: | 4-[(6-CHLORO-2-NAPHTHALENYL)SULFONYL]-1-[[1-(4-PYRIDINYL)-4-PIPERIDINYL] METHYL]-2-PIPERAZINECARBOXYLIC ACID, CALCIUM ION, coagulation Factor Xa | | Authors: | Shiromizu, I, Matsusue, T. | | Deposit date: | 2001-07-23 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Factor Xa Specific Inhibitor that Induces the Novel Binding Model in Complex with Human Fxa
To be Published
|
|
2PV1
 
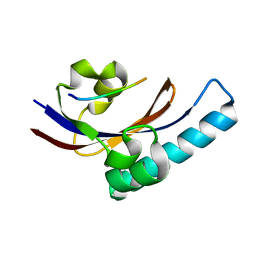 | |
5A8D
 
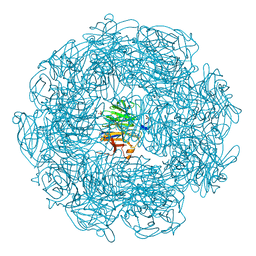 | | The high resolution structure of a novel alpha-L-arabinofuranosidase (CtGH43) from Clostridium thermocellum ATCC 27405 | | Descriptor: | ACETATE ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Goyal, A, Ahmed, S, Sharma, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-07-14 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular determinants of substrate specificity revealed by the structure of Clostridium thermocellum arabinofuranosidase 43A from glycosyl hydrolase family 43 subfamily 16.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7KFP
 
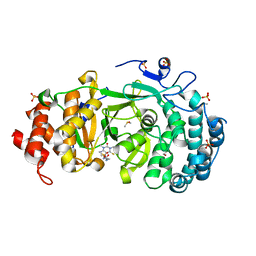 | | Structure of human PARG complexed with PARG-119 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{[2-(1,3-dimethyl-2-oxo-6-sulfanylidene-1,2,3,6-tetrahydro-7H-purin-7-yl)ethyl]carbamoyl}methanesulfonamide, ... | | Authors: | Brosey, C.A, Bommagani, S, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG7
 
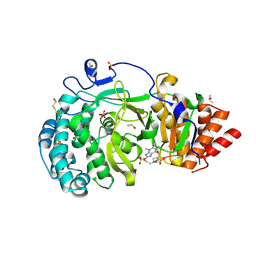 | | Structure of human PARG complexed with PARG-292 | | Descriptor: | 8-{[2-(1,1-dioxo-1lambda~6~,4-thiazinan-4-yl)ethyl]sulfanyl}-1,3-dimethyl-3,7-dihydro-1H-purine-2,6-dione, DIMETHYL SULFOXIDE, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
2PUP
 
 | |
2BDZ
 
 | | Mexicain from Jacaratia mexicana | | Descriptor: | Mexicain, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Gavira, J.A, Oliver-Salvador, M.C, Gonzalez-Ramirez, L.A, Soriano-Garcia, M, Garcia-Ruiz, J.M. | | Deposit date: | 2005-10-21 | | Release date: | 2006-10-03 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the mexicain-E-64 complex and comparison with other cysteine proteases of the papain family.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1J6P
 
 | |
1U4G
 
 | | Elastase of Pseudomonas aeruginosa with an inhibitor | | Descriptor: | CALCIUM ION, Elastase, N-(1-CARBOXY-3-PHENYLPROPYL)PHENYLALANYL-ALPHA-ASPARAGINE, ... | | Authors: | Bitto, E, McKay, D.B. | | Deposit date: | 2004-07-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Elastase of Pseudomonas aeruginosa with an inhibitor
To be Published, 2004
|
|
4IBQ
 
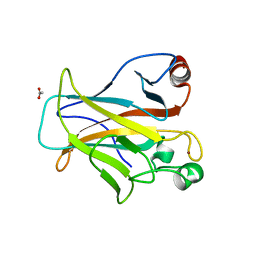 | | Human p53 core domain with hot spot mutation R273C | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Eldar, A, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
3UUE
 
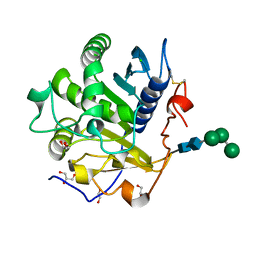 | | Crystal structure of mono- and diacylglycerol lipase from Malassezia globosa | | Descriptor: | CHLORIDE ION, GLYCEROL, LIP1, ... | | Authors: | Xu, T, Xu, J, Hou, S, Liu, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity.
J.Struct.Biol., 178, 2012
|
|
5SQA
 
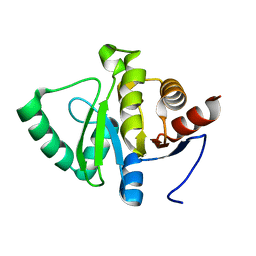 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894395 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7EXJ
 
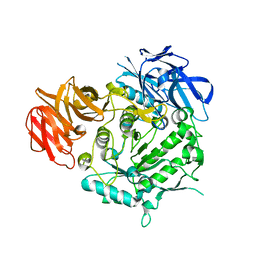 | | Crystal structure of alkaline alpha-galctosidase D383A mutant from Arabidopsis thaliana complexed with Raffinose | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
