3F6Q
 
 | | Crystal structure of integrin-linked kinase ankyrin repeat domain in complex with PINCH1 LIM1 domain | | Descriptor: | IODIDE ION, Integrin-linked protein kinase, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Chiswell, B.P, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2008-11-06 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis of integrin-linked kinase-PINCH interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7D83
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-(2-(3-cyclohexylureido)-3,6-dimethyl-5-(5-methylchroman-6-yl)pyridin-4-yl)acetic acid | | Descriptor: | (2S)-2-[2-(cyclohexylcarbamoylamino)-3,6-dimethyl-5-(5-methyl-3,4-dihydro-2H-chromen-6-yl)pyridin-4-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase, SULFATE ION | | Authors: | Sugiyama, S, Sekiguchi, Y. | | Deposit date: | 2020-10-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of novel HIV-1 integrase-LEDGF/p75 allosteric inhibitors based on a pyridine scaffold forming an intramolecular hydrogen bond.
Bioorg.Med.Chem.Lett., 33, 2020
|
|
2IVU
 
 | |
7DEL
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 3-[4-[2-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]ethynyl]phenyl]propan-1-ol, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Tanaka-Yamamoto, N. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
7DEM
 
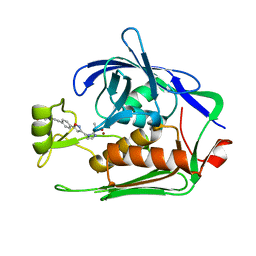 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 5-[4-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]phenyl]pent-4-yn-1-ol, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Matsuda, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
5O9Z
 
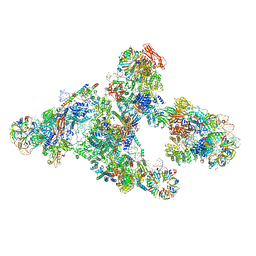 | |
5O8O
 
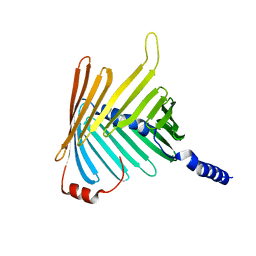 | | N. crassa Tom40 model based on cryo-EM structure of the TOM core complex at 6.8 A | | Descriptor: | Mitochondrial import receptor subunit tom40 | | Authors: | Bausewein, T, Mills, D.J, Nussberger, S, Nitschke, B, Kuehlbrandt, W. | | Deposit date: | 2017-06-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM Structure of the TOM Core Complex from Neurospora crassa.
Cell, 170, 2017
|
|
7D6T
 
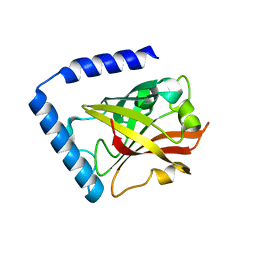 | |
7BMT
 
 | | HEWL in sodium chloride | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
2JIH
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (complex-form) | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAMTS-1, CADMIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains.
J. Mol. Biol., 373, 2007
|
|
2B90
 
 | | Crystal structure of the interleukin-4 variant T13DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B91
 
 | | Crystal structure of the interleukin-4 variant F82DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8Z
 
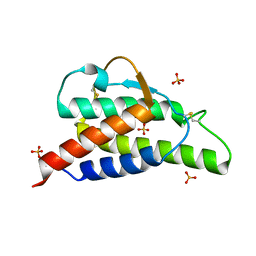 | | Crystal structure of the interleukin-4 variant R85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2LKB
 
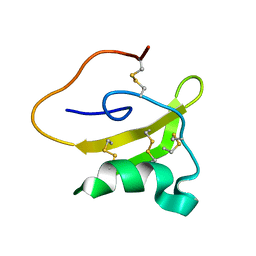 | | Evolutionary diversification of Mesobuthus alpha-scorpion toxins affecting sodium channels | | Descriptor: | Neurotoxin MeuNaTx-5 | | Authors: | Zhu, S, Peigneur, S, Gao, B, Lu, X, Cao, C, Tytgat, J. | | Deposit date: | 2011-10-09 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Evolutionary diversification of Mesobuthus {alpha}-scorpion toxins affecting sodium channels
Mol.Cell Proteomics, 2011
|
|
2BZS
 
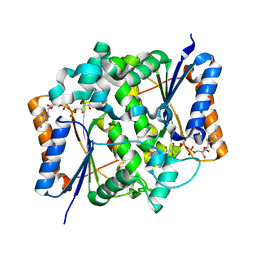 | | Binding of anti-cancer prodrug CB1954 to the activating enzyme NQO2 revealed by the crystal structure of their complex. | | Descriptor: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, NRH DEHYDROGENASE [QUINONE] 2, ... | | Authors: | Abu Khader, M.M, Heap, J.T, De Matteis, C, Kellam, B, Doughty, S.W, Minton, N, Paoli, M. | | Deposit date: | 2005-08-22 | | Release date: | 2005-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of the Anticancer Prodrug Cb1954 to the Activating Enzyme Nqo2 Revealed by the Crystal Structure of Their Complex.
J.Med.Chem., 48, 2005
|
|
7ECT
 
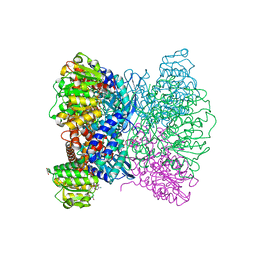 | | Crystal Structure of Aspergillus terreus Glutamate Dehydrogenase (AtGDH) Complexed With Tartrate and NADPH | | Descriptor: | GLYCEROL, Glutamate dehydrogenase, L(+)-TARTARIC ACID, ... | | Authors: | Godsora, B.K.J, Prakash, P, Punekar, N.S, Bhaumik, P. | | Deposit date: | 2021-03-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular insights into the inhibition of glutamate dehydrogenase by the dicarboxylic acid metabolites.
Proteins, 90, 2022
|
|
7ECR
 
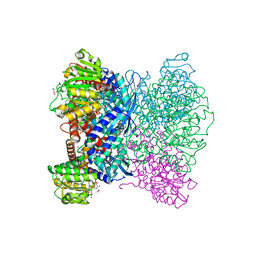 | | Crystal Structure of Aspergillus terreus Glutamate Dehydrogenase (AtGDH) Complexed With Succinate and ADP-ribose | | Descriptor: | GLYCEROL, Glutamate dehydrogenase, SUCCINIC ACID, ... | | Authors: | Godsora, B.K.J, Prakash, P, Punekar, N.S, Bhaumik, P. | | Deposit date: | 2021-03-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular insights into the inhibition of glutamate dehydrogenase by the dicarboxylic acid metabolites.
Proteins, 90, 2022
|
|
7ECS
 
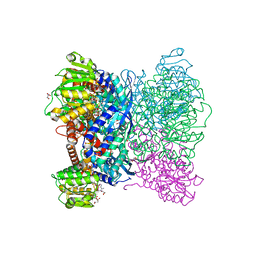 | | Crystal Structure of Aspergillus terreus Glutamate Dehydrogenase (AtGDH) Complexed With Malonate and NADPH | | Descriptor: | GLYCEROL, Glutamate dehydrogenase, MALONATE ION, ... | | Authors: | Godsora, B.K.J, Prakash, P, Punekar, N.S, Bhaumik, P. | | Deposit date: | 2021-03-13 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular insights into the inhibition of glutamate dehydrogenase by the dicarboxylic acid metabolites.
Proteins, 90, 2022
|
|
7EGU
 
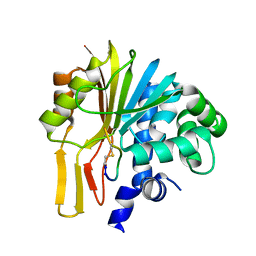 | | Structure of human NNMT in complex with macrocyclic peptide X | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide X | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
7DCF
 
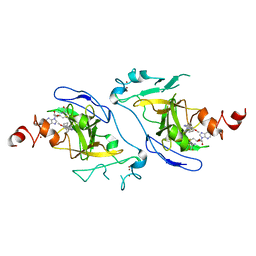 | | Crystal structure of EHMT2 SET domain in complex with compound 10 | | Descriptor: | 5'-methoxy-6'-(1-methyl-2,3,4,7-tetrahydroazepin-5-yl)spiro[cyclobutane-1,3'-indole]-2'-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Suzuki, M, Katayama, K. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of DS79932728: A Potent, Orally Available G9a/GLP Inhibitor for Treating beta-Thalassemia and Sickle Cell Disease.
Acs Med.Chem.Lett., 12, 2021
|
|
3G0V
 
 | |
3FYD
 
 | |
2JKC
 
 | | Crystal Structure of E346D of Tryptophan 7-Halogenase (PrnA) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Zhu, X, Naismith, J.H. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights into the mechanism of enzymatic chlorination of tryptophan.
Angew. Chem. Int. Ed. Engl., 47, 2008
|
|
5O0H
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 2-(4-Chloro-3-nitrobenzoyl)benzoic acid (Fragment 6) | | Descriptor: | 2-(4-chloranyl-3-nitro-phenyl)carbonylbenzoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
7EKZ
 
 | | Structural and functional insights into Hydra Actinoporin-Like Toxin 1 (HALT-1) | | Descriptor: | FORMIC ACID, GLYCEROL, HALT-1 | | Authors: | Ker, D.S, Sha, X.H, Jonet, M.A, Hwang, J.S, Ng, C.L. | | Deposit date: | 2021-04-07 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and functional analysis of Hydra Actinoporin-Like Toxin 1 (HALT-1).
Sci Rep, 11, 2021
|
|
