1RKD
 
 | | E. COLI RIBOKINASE COMPLEXED WITH RIBOSE AND ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOKINASE, ... | | Authors: | Sigrell, J.A, Cameron, A.D, Jones, T.A, Mowbray, S.L. | | Deposit date: | 1997-11-29 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 A resolution: insights into a new family of kinase structures.
Structure, 6, 1998
|
|
2BRM
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | 3-AMINO-3-BENZYL-[4.3.0]BICYCLO-1,6-DIAZANONAN-2-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
3BL6
 
 | | Crystal structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Siu, K.K.W, Lee, J.E, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1V41
 
 | | Crystal structure of human PNP complexed with 8-Azaguanine | | Descriptor: | 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Dos Santos, D.M, Canduri, F, Silva, R.G, Mendes, M.A, Basso, L.A, Palma, M.S, De Azevedo Jr, W.F, Santos, D.S. | | Deposit date: | 2003-11-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of human PNP complexed with ligands.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BME
 
 | | high resolution structure of GppNHp-bound human Rab4a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Scheidig, A.J, Huber, S.K. | | Deposit date: | 2005-03-13 | | Release date: | 2005-04-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | High Resolution Crystal Structures of Human Rab4A in its Active and Inactive Conformations.
FEBS Lett., 579, 2005
|
|
5PAV
 
 | | Crystal Structure of Factor VIIa in complex with N-(6-aminopyridin-3-yl)-5-hydroxy-1-phenylpyrazole-4-carboxamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII heavy chain, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
4G20
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}-2'-methyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
2BRC
 
 | | Structure of a Hsp90 Inhibitor bound to the N-terminus of Yeast Hsp90. | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
4JTZ
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 4 | | Descriptor: | 3-{[4-oxo-1-(2,4,6-trifluorobenzyl)-1,4-dihydroquinazolin-6-yl]oxy}-N-(pyridin-3-yl)-2-(trifluoromethyl)benzamide, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
4JUA
 
 | |
1RHJ
 
 | |
4BF4
 
 | |
1AQB
 
 | | RETINOL-BINDING PROTEIN (RBP) FROM PIG PLASMA | | Descriptor: | CADMIUM ION, RETINOL, RETINOL-BINDING PROTEIN | | Authors: | Zanotti, G, Panzalorto, M, Marcato, A, Malpeli, G, Folli, C, Berni, R. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pig plasma retinol-binding protein at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
4AV1
 
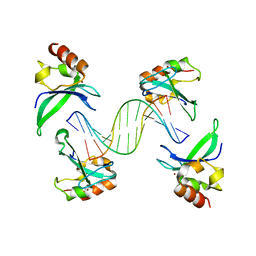 | | Crystal structure of the human PARP-1 DNA binding domain in complex with DNA | | Descriptor: | 5'-D(*AP*AP*GP*TP*GP*TP*TP*GP*CP*AP*TP*TP)-3', 5'-D(*TP*AP*AP*TP*GP*CP*AP*AP*CP*AP*CP*TP)-3', POLY [ADP-RIBOSE] POLYMERASE 1, ... | | Authors: | Ali, A.A.E, Timinszky, G, Arribas-Bosacoma, R, Kozlowski, M, Hassa, P.O, Hassler, M, Ladurner, A.G, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Zinc-Finger Domains of Parp1 Cooperate to Recognise DNA Strand-Breaks
Nat.Struct.Mol.Biol., 19, 2012
|
|
5V7Q
 
 | | Cryo-EM structure of the large ribosomal subunit from Mycobacterium tuberculosis bound with a potent linezolid analog | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Zhang, J. | | Deposit date: | 2017-03-20 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
4JZV
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4Q7X
 
 | | Neutrophil serine protease 4 (PRSS57) apo form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Serine protease 57, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
1PMO
 
 | | Crystal structure of Escherichia coli GadB (neutral pH) | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate decarboxylase beta | | Authors: | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | Deposit date: | 2003-06-11 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
1PQN
 
 | | dominant negative human hDim1 (hDim1 1-128) | | Descriptor: | Spliceosomal U5 snRNP-specific 15 kDa protein | | Authors: | Zhang, Y.Z, Cheng, H, Gould, K.L, Golemis, E.A, Roder, H. | | Deposit date: | 2003-06-18 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, stability and function of hDim1 investigated by NMR, circular
dichroism and mutational analysis
Biochemistry, 42, 2003
|
|
3MVX
 
 | | X-ray structure of the reduced NikA/1 hybrid, NikA/1-Red | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2-[2-[carboxymethyl(phenylmethyl)amino]ethyl-[(2-hydroxyphenyl)methyl]amino]ethanoic acid, ... | | Authors: | Cavazza, C, Bochot, C, Rousselot-Pailley, P, Carpentier, P, Cherrier, M.V, Martin, L, Marchi-Delapierre, C, Fontecilla-Camps, J.C, Menage, S. | | Deposit date: | 2010-05-05 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of the reaction of aromatic C-H with O(2) catalysed by a protein-bound iron complex
NAT.CHEM., 2, 2010
|
|
2XJX
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, Onalespib | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
3W11
 
 | |
4FV3
 
 | |
7JWU
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 1-methyl-5-phenyl-6-{[(1R)-1-(pyridin-2-yl)ethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
