3QA3
 
 | | Crystal Structure of A-domain in complex with antibody | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy chain, Antibody Light chain, ... | | Authors: | Mahalingam, B, Xiong, J.P, Arnaout, M.A. | | Deposit date: | 2011-01-10 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stable Coordination of the Inhibitory Ca2+ Ion at the Metal Ion-Dependent Adhesion Site in Integrin CD11b/CD18 by an Antibody-Derived Ligand Aspartate: Implications for Integrin Regulation and Structure-Based Drug Design.
J.Immunol., 187, 2011
|
|
1K1O
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, {[(1R)-2-((2S)-2-{[(3-{[AMINO(IMINO)METHYL]AMINO}PROPYL)AMINO]CARBONYL}PIPERIDINYL)-1-(CYCLOHEXYLMETHYL)-2-OXOETHYL]AMINO}ACETIC ACID | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
1K22
 
 | | HUMAN THROMBIN-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-2, Prothrombin, ... | | Authors: | Stubbs, M.T, Musil, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
6GHT
 
 | | HtxB D206A protein variant from Pseudomonas stutzeri in complex with hypophosphite to 1.12 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2018-05-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
6D26
 
 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant | | Descriptor: | OLEIC ACID, Prostaglandin D2 receptor 2, Endolysin chimera, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
2VFE
 
 | | Crystal structure of F96S mutant of Plasmodium falciparum triosephosphate isomerase with 3- phosphoglycerate bound at the dimer interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCEROL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7TX5
 
 | |
7TX3
 
 | |
7TX4
 
 | |
6C3M
 
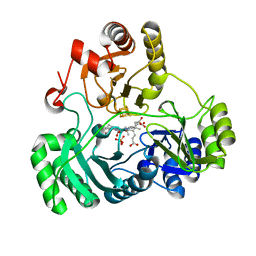 | | Wild type structure of SiRHP | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stroupe, M.E. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The role of extended Fe4S4cluster ligands in mediating sulfite reductase hemoprotein activity.
Biochim. Biophys. Acta, 1866, 2018
|
|
6C3X
 
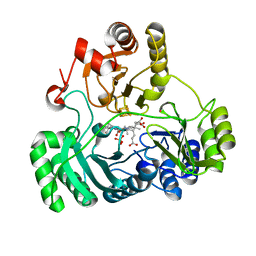 | | Wild type structure of SiRHP | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stroupe, M.E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.542 Å) | | Cite: | The role of extended Fe4S4cluster ligands in mediating sulfite reductase hemoprotein activity.
Biochim. Biophys. Acta, 1866, 2018
|
|
3VJS
 
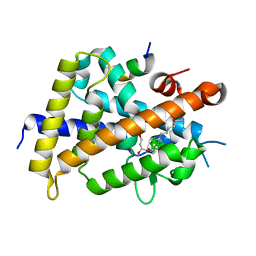 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(S)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
4ZSH
 
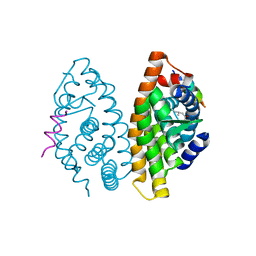 | | RXR LBD in complex with 9-cis-13,14-dihydroretinoic acid | | Descriptor: | (5S,6S,9R,13R)-2,3-didehydro-5,6,7,8,9,10,11,12,13,14-decahydroretinoic acid, NCoA2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Rochel, N, Krezel, W, Ruhl, R. | | Deposit date: | 2015-05-13 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 9-cis-13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice.
Plos Genet., 11, 2015
|
|
6D27
 
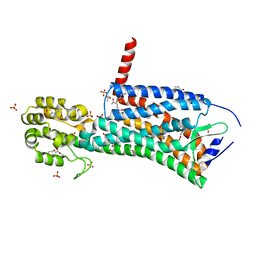 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
3RTT
 
 | | Human MMP-12 catalytic domain in complex with*(R)-N*-Hydroxy-1-(phenethylsulfonyl)pyrrolidine-2-carboxamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-1-[(2-phenylethyl)sulfonyl]-D-prolinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
4PP8
 
 | |
3RTS
 
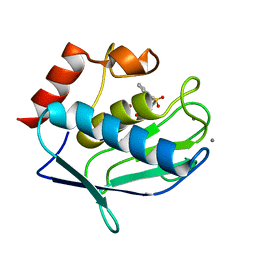 | | Human MMP-12 catalytic domain in complex with*N*-Hydroxy-2-(2-phenylethylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-[(2-phenylethyl)sulfonyl]glycinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
7LIR
 
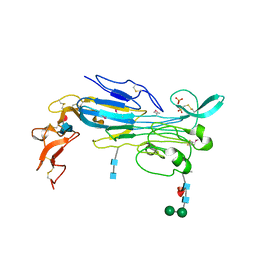 | | Structure of the invertebrate ALK GRD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALK tyrosine kinase receptor homolog scd-2, ... | | Authors: | Stayrook, S, Li, T, Klein, D.E. | | Deposit date: | 2021-01-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
1K1P
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, TRYPSIN, [((1R)-2-{(2S)-2-[({4-[AMINO(IMINO)METHYL]BENZYL}AMINO)CARBONYL]AZETIDINYL}-1-CYCLOHEXYL-2-OXOETHYL)AMINO]ACETIC ACID | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
1K1J
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, N-ALPHA-(2-NAPHTHYLSULFONYL)-N(3-AMIDINO-L-PHENYLALANINYL)ISOPIPECOLINIC ACID METHYL ESTER, SULFATE ION, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
1K1M
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, N-ALPHA-(2-NAPHTHYLSULFONYL)-N(3-AMIDINO-L-PHENYLALANINYL)-4-ACETYL-PIPERAZINE, SULFATE ION, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
3VJT
 
 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(R)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
4PUU
 
 | |
2VFF
 
 | | Crystal structure of the F96H mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFI
 
 | | Crystal structure of the Plasmodium falciparum triosephosphate isomerase in the loop closed state with 3-phosphoglycerate bound at the active site and interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
