5RXP
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2856434944 | | Descriptor: | DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1, ~{N}-(4-fluorophenyl)-4-methyl-piperazine-1-carboxamide | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RY1
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2940170964 | | Descriptor: | (3R)-1-methylpiperidin-3-yl furan-2-carboxylate, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
1OS6
 
 | | Cytochrome c7 (PpcA) from Geobacter sulfurreducens | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, HEME C, PpcA, ... | | Authors: | Pokkuluri, P.R, Londer, Y.Y, Duke, N.E.C, Long, W.C, Schiffer, M. | | Deposit date: | 2003-03-18 | | Release date: | 2004-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Family of cytochrome c7-type proteins from Geobacter sulfurreducens: structure of one cytochrome c7 at 1.45 A resolution.
Biochemistry, 43, 2004
|
|
3W53
 
 | |
5RYO
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z136583524 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-N-(pyridin-4-yl)furan-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5HMJ
 
 | |
4R5E
 
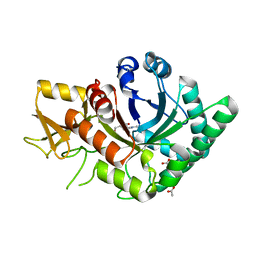 | | Crystal Structure of Family GH18 Chitinase from Cycas revoluta a Complex with Allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ACETATE ION, ALLOSAMIZOLINE, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2014-08-21 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures and inhibitor binding properties of plant class V chitinases: the cycad enzyme exhibits unique structural and functional features.
Plant J., 82, 2015
|
|
5RY6
 
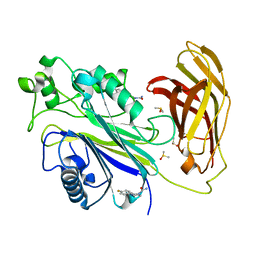 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z198194394 | | Descriptor: | 4-(4-fluorophenyl)piperazine-1-carboxamide, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
3PJX
 
 | |
5RYR
 
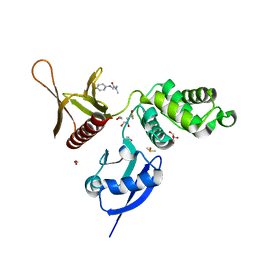 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z104584152 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZX
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434814 | | Descriptor: | 1,2-ETHANEDIOL, 3-(diethylamino)phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZV
 
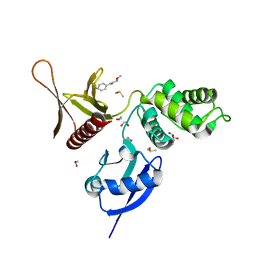 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z31504642 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-phenylacetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
4IWQ
 
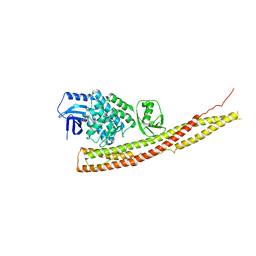 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(morpholin-4-ylmethyl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
2LK9
 
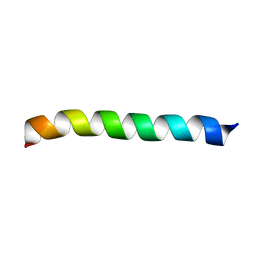 | | Structure of BST-2/Tetherin Transmembrane Domain | | Descriptor: | Bone marrow stromal antigen 2 | | Authors: | Skasko, M, Wang, Y, Tian, Y, Tokarev, A, Munguia, J, Ruiz, A, Stephens, E, Opella, S, Guatelli, J. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 Vpu Protein Antagonizes Innate Restriction Factor BST-2 via Lipid-embedded Helix-Helix Interactions.
J.Biol.Chem., 287, 2012
|
|
4IXC
 
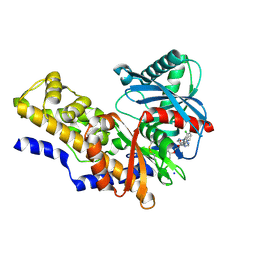 | |
5T9W
 
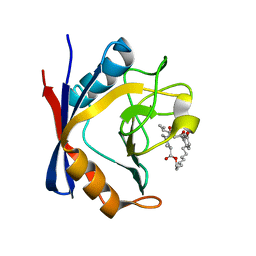 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 5) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 3-[(3-hydroxyphenyl)methyl]-6-(propan-2-yl)-19-oxa-1,4,7,25-tetraazabicyclo[19.3.1]pentacosa-13,15-diene-2,5,8,20-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, P, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
4OAV
 
 | | Complete human RNase L in complex with 2-5A (5'-ppp heptamer), AMPPCP and RNA substrate. | | Descriptor: | (2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-hydroxy-2-({[(S)-hydroxy{[(2R,3S,4S)-4-hydroxy-2-(hydroxymethyl)tetrahydrofuran-3-yl]oxy}phosphoryl]oxy}methyl)tetrahydrofuran-3-yl dihydrogen phosphate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Han, Y, Donovan, J, Rath, S, Whitney, G, Chitrakar, A, Korennykh, A. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response.
Science, 343, 2014
|
|
6B31
 
 | | Structure of RORgt in complex with a novel inverse agonist 2 | | Descriptor: | (3S)-N~1~-(3-chloro-4-cyanophenyl)-N~5~-(1,3-diethyl-2,4-dioxo-1,2,3,4-tetrahydroquinazolin-6-yl)-3-methylpentanediamide, Nuclear receptor ROR-gamma | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Identification of novel quinazolinedione derivatives as ROR gamma t inverse agonist.
Bioorg. Med. Chem., 26, 2018
|
|
4GCY
 
 | | Structure of Mycobacterium tuberculosis dUTPase H21W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Toth, J, Vertessy, B.G, Leveles, I, Bendes, A. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | RAMD identification of substrate binding pathways to the active site of dUTPase
To be Published
|
|
2BLN
 
 | | N-terminal formyltransferase domain of ArnA in complex with N-5- formyltetrahydrofolate and UMP | | Descriptor: | ACETATE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PROTEIN YFBG, ... | | Authors: | Williams, G.J, Breazeale, S.D, Raetz, C.R.H, Naismith, J.H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and Function of Both Domains of Arna, a Dual Function Decarboxylase and a Formyltransferase, Involved in 4-Amino-4-Deoxy-L-Arabinose Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
3Q5U
 
 | |
5R95
 
 | | PanDDA analysis group deposition Form1 MAP kinase p38-alpha -- Fragment KCL093 in complex with MAP kinase p38-alpha | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-aminophenyl)pyrrole-2,5-dione, CHLORIDE ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
3Q6L
 
 | | Crystal Structure of Human Adipocyte Fatty Acid Binding Protein (FABP4) at 1.4 Ang. Resolution | | Descriptor: | Fatty acid-binding protein, adipocyte | | Authors: | Prasad, G.S, Carney, D, Small, V, Sankaran, B, Zwart, P.H. | | Deposit date: | 2011-01-02 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Human Adipocyte Fatty Acid Binding Protein (FABP4) at 1.4 Ang. Resolution
To be Published
|
|
3GXN
 
 | | Crystal structure of apo alpha-galactosidase A at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, SULFATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
2ZJY
 
 | | Structure of the K349P mutant of Gi alpha 1 subunit bound to ALF4 and GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Morikawa, T, Muroya, A, Sugio, S, Wakamatsu, K, Kohno, T. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How GPCRs activate G proteins: Structural changes form C-terminal tail to GDP binding pocket
To be Published
|
|
