5KQJ
 
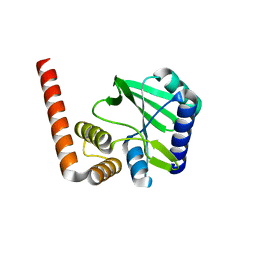 | |
5LO4
 
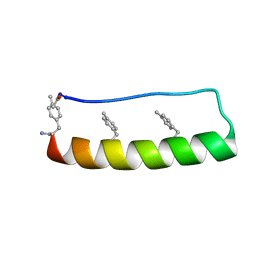 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPa-CH3 | | Authors: | Baker, E.G, Williams, C, Hudson, K.L, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5NPA
 
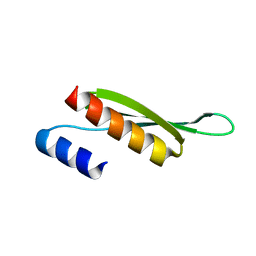 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | Descriptor: | Loquacious | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5NPG
 
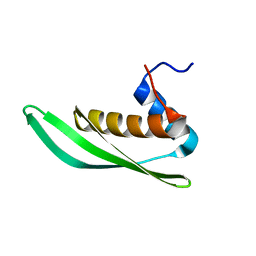 | | Solution structure of Drosophila melanogaster Loquacious dsRBD1 | | Descriptor: | Loquacious, isoform F | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Hartlmuller, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
6IR0
 
 | | Zinc finger domain of the human DTX protein | | Descriptor: | Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Miyamoto, K. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc finger domain of the human DTX protein adopts a unique RING fold.
Protein Sci., 28, 2019
|
|
5LO2
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaTyr | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LO3
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaOMe | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
1FHO
 
 | | Solution Structure of the PH Domain from the C. Elegans Muscle Protein UNC-89 | | Descriptor: | UNC-89 | | Authors: | Blomberg, N, Baraldi, E, Sattler, M, Saraste, M, Nilges, M. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a PH domain from the C. elegans muscle protein UNC-89 suggests a novel function.
Structure Fold.Des., 8, 2000
|
|
2YMJ
 
 | |
5NWM
 
 | |
1PJV
 
 | | Cobatoxin 1 from Centruroides noxius Scorpion venom: Chemical Synthesis, 3-D Structure in Solution, Pharmacology and Docking on K+ channels | | Descriptor: | Cobatoxin 1 | | Authors: | Mosbah, A, Jouirou, B, Visan, V, Grissmer, S, El Ayeb, M, Rochat, H, De Waard, M, Mabrouk, K, Sabatier, J.M. | | Deposit date: | 2003-06-03 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Cobatoxin 1 from Centruroides noxius scorpion venom: chemical synthesis, three-dimensional structure in solution, pharmacology and docking on K+ channels.
Biochem.J., 377, 2004
|
|
1BZB
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | PROTEIN (CALCITONIN), alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-10-27 | | Release date: | 1998-11-11 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
7QDQ
 
 | | Crystal Structure of HDM2 in complex with Caylin-1 | | Descriptor: | CHLORIDE ION, Caylin-1, DIMETHYL SULFOXIDE, ... | | Authors: | Finke, A.D, Walti, M.A, Marsh, M.E, Orts, J. | | Deposit date: | 2021-11-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Elucidation of a nutlin-derivative-HDM2 complex structure at the interaction site by NMR molecular replacement: A straightforward derivation
J Magn Reson Open, 10-11, 2022
|
|
1NMW
 
 | | Solution structure of the PPIase domain of human Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | Authors: | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
1NYO
 
 | | Solution structure of the antigenic TB protein MPT70/MPB70 | | Descriptor: | Immunogenic protein MPT70 | | Authors: | Bloemink, M.J, Dentten, E, Hewinson, R.G, Williamson, R.A, Carr, M.D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-02-13 | | Release date: | 2003-08-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Mycobacterium tuberculosis complex protein MPB70: from tuberculosis pathogenesis to inherited human corneal desease
J.Biol.Chem., 278, 2003
|
|
7R3M
 
 | |
6EKA
 
 | | Solid-state MAS NMR structure of the HELLF prion amyloid fibrils | | Descriptor: | Podospora anserina S mat+ genomic DNA chromosome 3, supercontig 2 | | Authors: | Martinez, D, Daskalov, A, Andreas, L, Bardiaux, B, Coustou, V, Stanek, J, Berbon, M, Noubhani, M, Kauffmann, B, Wall, J.S, Pintacuda, G, Saupe, S.J, Habenstein, B, Loquet, A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structural and molecular basis of cross-seeding barriers in amyloids
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5Y95
 
 | | Haddock model of mSIN3B PAH1 domain | | Descriptor: | Paired amphipathic helix protein Sin3b, propan-2-yl (3R,6S,9aS)-3-ethyl-8-(3-methylbutyl)-6-(2-methylsulfanylethyl)-4,7-bis(oxidanylidene)-9,9a-dihydro-6H-pyrazino[2,1-c][1,2,4]oxadiazine-1-carboxylate | | Authors: | Kurita, J, Hirao, Y, Nishimura, Y. | | Deposit date: | 2017-08-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A mimetic of the mSin3-binding helix of NRSF/REST ameliorates abnormal pain behavior in chronic pain models.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7SAW
 
 | | Mu-conotoxin KIIIA isomer 2 | | Descriptor: | Mu-conotoxin KIIIA | | Authors: | Schroeder, C.I, Tran, H.N.T. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the inhibition of human voltage-gated sodium channels by mu-conotoxin KIIIA disulfide isomers.
J.Biol.Chem., 298, 2022
|
|
5Y0I
 
 | | Solution structure of arenicin-3 derivative N1 | | Descriptor: | NZ17074(N1) | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
5YJ4
 
 | |
5Y0H
 
 | | Solution structure of arenicin-3 derivative N6 | | Descriptor: | N6 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
6HIR
 
 | |
7GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
1N02
 
 | |
