3NCY
 
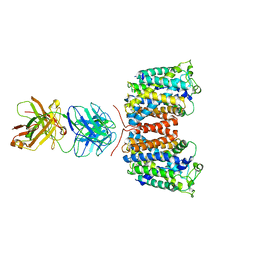 | | X-ray crystal structure of an arginine agmatine antiporter (AdiC) in complex with a Fab fragment | | Descriptor: | AdiC, Fab Heavy chain, Fab Light chain | | Authors: | Fang, Y, Jayaram, H, Shane, T, Komalkova-Partensky, L, Wu, F, Williams, C, Xiong, Y, Miller, C. | | Deposit date: | 2010-06-06 | | Release date: | 2010-08-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a prokaryotic virtual proton pump at 3.2 A resolution.
Nature, 460, 2009
|
|
4QL0
 
 | | Crystal Structure Analysis of the Membrane Transporter FhaC (double mutant V169T, I176N) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Filamentous hemagglutinin transporter protein FhaC, HEXAETHYLENE GLYCOL, ... | | Authors: | Maier, T, Clantin, B, Gruss, F, Dewitte, F, Delattre, A.S, Jacob-Dubuisson, F, Hiller, S, Villeret, V. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conserved Omp85 lid-lock structure and substrate recognition in FhaC
Nat Commun, 6, 2015
|
|
2HEG
 
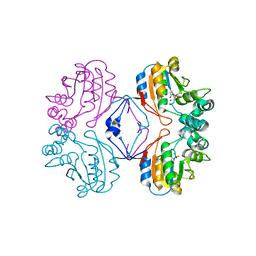 | | Phospho-Aspartyl Intermediate Analogue of Apha class B acid phosphatase/phosphotransferase | | Descriptor: | Class B acid phosphatase, MAGNESIUM ION | | Authors: | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | Deposit date: | 2006-06-21 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
|
|
2H4W
 
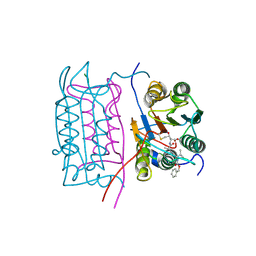 | |
1RFV
 
 | |
2O56
 
 | | Crystal Structure of a Member of the Enolase Superfamily from Salmonella Typhimurium | | Descriptor: | MAGNESIUM ION, Putative mandelate racemase | | Authors: | Patskovsky, Y, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Putative Enolase from Salmonella Typhimurium Lt2
To be Published
|
|
4C4V
 
 | |
4N7X
 
 | |
4EQB
 
 | | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein PotD from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES.
TO BE PUBLISHED
|
|
5A2N
 
 | |
2PL5
 
 | | Crystal Structure of Homoserine O-acetyltransferase from Leptospira interrogans | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase | | Authors: | Liu, L, Wang, M, Wang, Y, Wei, Z, Xu, H, Gong, W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of homoserine O-acetyltransferase from Leptospira interrogans
Biochem.Biophys.Res.Commun., 363, 2007
|
|
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
2P8B
 
 | | Crystal structure of N-succinyl Arg/Lys racemase from Bacillus cereus ATCC 14579 complexed with N-succinyl Lys. | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, N-SUCCINYL LYSINE | | Authors: | Fedorov, A.A, Song, L, Fedorov, E.V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Prediction and assignment of function for a divergent N-succinyl amino acid racemase.
Nat.Chem.Biol., 3, 2007
|
|
4KSY
 
 | |
2AZ5
 
 | | Crystal Structure of TNF-alpha with a small molecule inhibitor | | Descriptor: | 6,7-DIMETHYL-3-[(METHYL{2-[METHYL({1-[3-(TRIFLUOROMETHYL)PHENYL]-1H-INDOL-3-YL}METHYL)AMINO]ETHYL}AMINO)METHYL]-4H-CHROMEN-4-ONE, Tumor necrosis factor (TNF-alpha) (Tumor necrosis factor ligand superfamily member 2) (TNF-a) (Cachectin) [Contains: Tumor necrosis factor, membrane form; Tumor necrosis factor, ... | | Authors: | He, M.M. | | Deposit date: | 2005-09-09 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-molecule inhibition of TNF-alpha.
Science, 310, 2005
|
|
4B8R
 
 | | Crystal Structure of Thermococcus litoralis ADP-dependent Glucokinase (GK) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADP-DEPENDENT GLUCOKINASE, ... | | Authors: | Herrera-Morande, A, Rivas-Pardo, J.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure, Saxs and Kinetic Mechanism of Hyperthermophilic Adp-Dependent Glucokinase from Thermococcus Litoralis Reveal a Conserved Mechanism for Catalysis.
Plos One, 8, 2013
|
|
1UMA
 
 | | ALPHA-THROMBIN (HIRUGEN) COMPLEXED WITH NA-(N,N-DIMETHYLCARBAMOYL)-ALPHA-AZALYSINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUDIN I, ... | | Authors: | Nardini, M, Pesce, A, Rizzi, M, Casale, E, Ferraccioli, R, Balliano, G, Milla, P, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1996-03-26 | | Release date: | 1996-11-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the active site titrant N alpha-(N,N-dimethylcarbamoyl)-alpha-azalysine p-nitrophenyl ester: a comparative kinetic and X-ray crystallographic study.
J.Mol.Biol., 258, 1996
|
|
4PO6
 
 | |
4N7W
 
 | | Crystal Structure of the sodium bile acid symporter from Yersinia frederiksenii | | Descriptor: | CITRIC ACID, Transporter, sodium/bile acid symporter family, ... | | Authors: | Zhou, X, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of the alternating-access mechanism in a bile acid transporter.
Nature, 505, 2013
|
|
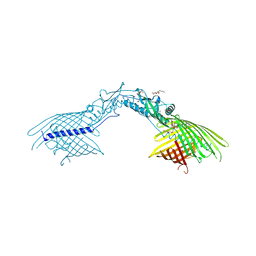
1HNG
 
 | | CRYSTAL STRUCTURE AT 2.8 ANGSTROMS RESOLUTION OF A SOLUBLE FORM OF THE CELL ADHESION MOLECULE CD2 | | Descriptor: | CD2 | | Authors: | Jones, E.Y, Davis, S.J, Williams, A.F, Harlos, K, Stuart, D.I. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure at 2.8 A resolution of a soluble form of the cell adhesion molecule CD2.
Nature, 360, 1992
|
|
4RXM
 
 | | Crystal structure of periplasmic ABC transporter solute binding protein A7JW62 from Mannheimia haemolytica PHL213, Target EFI-511105, in complex with Myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of solute binding sugar transporter A7JW62 from Mannheimia haemolytica, Target EFI-511105.
To be Published
|
|
2XH4
 
 | |
2KN6
 
 | |
5D4P
 
 | | Structure of CPII bound to ADP and bicarbonate, from Thiomonas intermedia K12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, Putative Nitrogen regulatory protein P-II GlnB | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
2XH7
 
 | |
