6T80
 
 | |
6Q0T
 
 | | Structure of a MAPK pathway complex | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Park, E, Rawson, S, Jeon, H, Eck, M.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-09 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes.
Nature, 575, 2019
|
|
5NAS
 
 | |
8CJ4
 
 | | Crystal structure of ClpP from Staphylococcus epidermidis, tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Alves Franca, B, Rohde, H, Betzel, C. | | Deposit date: | 2023-02-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the dynamic modulation of bacterial ClpP function and oligomerization by peptidomimetic boronate compounds.
Sci Rep, 14, 2024
|
|
6QIU
 
 | | Crystal structure of 14-3-3 sigma in complex with Ataxin-1 Ser776 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Ataxin-1 phosphopeptide, CHLORIDE ION, ... | | Authors: | Leysen, S, Milroy, L.G, Davis, J.M, Brunsveld, L, Ottmann, C. | | Deposit date: | 2019-01-21 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural insights into the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1
To Be Published
|
|
2FS3
 
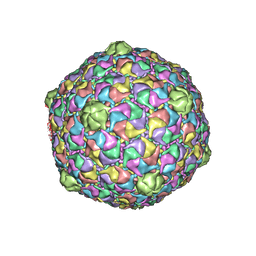 | | Bacteriophage HK97 K169Y Head I | | Descriptor: | Major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
2J57
 
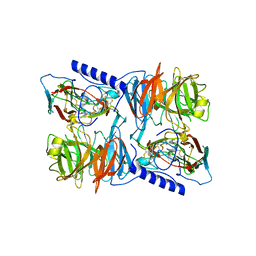 | | X-ray reduced Paraccocus denitrificans methylamine dehydrogenase N- quinol in complex with amicyanin. | | Descriptor: | AMICYANIN, COPPER (II) ION, METHYLAMINE DEHYDROGENASE HEAVY CHAIN, ... | | Authors: | Pearson, A.R, Pahl, R, Davidson, V.L, Wilmot, C.M. | | Deposit date: | 2006-09-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tracking X-Ray-Derived Redox Changes in Crystals of a Methylamine Dehydrogenase/Amicyanin Complex Using Single-Crystal Uv/Vis Microspectrophotometry.
J.Synchrotron Radiat., 14, 2007
|
|
2J56
 
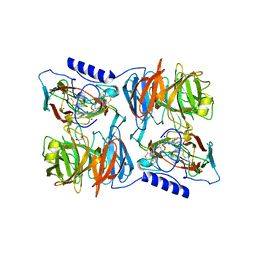 | | X-ray reduced Paraccocus denitrificans methylamine dehydrogenase N- semiquinone in complex with amicyanin. | | Descriptor: | AMICYANIN, COPPER (II) ION, GLYCEROL, ... | | Authors: | Pearson, A.R, Pahl, R, Davidson, V.L, Wilmot, C.M. | | Deposit date: | 2006-09-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tracking X-Ray-Derived Redox Changes in Crystals of a Methylamine Dehydrogenase/Amicyanin Complex Using Single-Crystal Uv/Vis Microspectrophotometry.
J.Synchrotron Radiat., 14, 2007
|
|
2J55
 
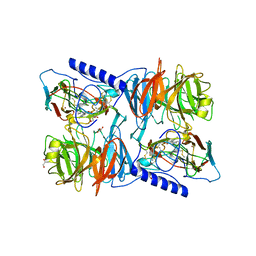 | | X-ray reduced Paraccocus denitrificans methylamine dehydrogenase O- quinone in complex with amicyanin. | | Descriptor: | AMICYANIN, COPPER (II) ION, GLYCEROL, ... | | Authors: | Pearson, A.R, Pahl, R, Davidson, V.L, Wilmot, C.M. | | Deposit date: | 2006-09-12 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Tracking X-Ray-Derived Redox Changes in Crystals of a Methylamine Dehydrogenase/Amicyanin Complex Using Single-Crystal Uv/Vis Microspectrophotometry.
J.Synchrotron Radiat., 14, 2007
|
|
1RWQ
 
 | | Human Dipeptidyl peptidase IV in complex with 5-aminomethyl-6-(2,4-dichloro-phenyl)-2-(3,5-dimethoxy-phenyl)-pyrimidin-4-ylamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(AMINOMETHYL)-6-(2,4-DICHLOROPHENYL)-2-(3,5-DIMETHOXYPHENYL)PYRIMIDIN-4-AMINE, Dipeptidyl peptidase IV | | Authors: | Hennig, M, Thoma, R, Stihle, M. | | Deposit date: | 2003-12-17 | | Release date: | 2004-12-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminomethylpyrimidines as novel DPP-IV inhibitors: a 10(5)-fold activity increase by optimization of aromatic substituents
Bioorg.Med.Chem.Lett., 14, 2004
|
|
6OHD
 
 | | P38 in complex with T-3220137 | | Descriptor: | 3-(3-tert-butyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-6-yl)-4-methyl-N-(1,2-oxazol-3-yl)benzamide, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]Pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 2.
Chemmedchem, 14, 2019
|
|
6OAR
 
 | |
7JM5
 
 | | Crystal structure of KDM4B in complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-07-31 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma.
Sci Transl Med, 14, 2022
|
|
8QYF
 
 | |
5F08
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium tuberculosis in complex with compound 14 at 1.92A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, ~{tert}-butyl ~{N}-methyl-~{N}-[[4-[4-(3-oxidanylidene-3-pyrrolidin-1-yl-propyl)piperidin-1-yl]phenyl]methyl]carbamate | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
2DE5
 
 | |
2DE6
 
 | |
7PCN
 
 | | BurG (holo) in complex with gonyenediol (14), trigonic acid (6) and DMS: Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R)-2-oxidanyl-2-(1-oxidanylcyclopropyl)ethanoic acid, (METHYLSULFANYL)METHANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
1ZA0
 
 | | X-ray structure of putative acyl-ACP desaturase DesA2 from Mycobacterium tuberculosis H37Rv | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, POSSIBLE ACYL-[ACYL-CARRIER PROTEIN] DESATURASE DESA2 (ACYL-[ACP] DESATURASE) (STEAROYL-ACP DESATURASE) | | Authors: | Dyer, H.D, Lyle, K.S, Rayment, I, Fox, B.G. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of putative acyl-ACP desaturase DesA2 from Mycobacterium tuberculosis H37Rv.
Protein Sci., 14, 2005
|
|
7Z6I
 
 | | Crystal structure of p38alpha C162S in complex with SB20358 and CAS 2094667-81-7 (behind catalytic site; Y35 in), P 21 21 21 | | Descriptor: | 4-[4-(4-fluorophenyl)-2-[4-[methyl(oxidanyl)-$l^{3}-sulfanyl]phenyl]-1~{H}-imidazol-5-yl]pyridine, Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)benzenesulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
7Z9T
 
 | | Crystal structure of p38alpha C162S in complex with ATPgS and CAS 2094667-81-7 (in catalytic site, Y35 out), P 1 21 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)benzenesulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-03-21 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
6Q9S
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 14 AT 2.4A: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[[6-chloranyl-3-[3-[(1~{S})-1-(2,4-dichlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-1~{H}-indol-2-yl]carbonylamino]-4-[4-(2-oxidanylidene-1,3-oxazinan-3-yl)piperidin-1-yl]benzoic acid, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
4BWW
 
 | | Crystal structure of spin labelled azurin T21R1. | | Descriptor: | AZURIN, COPPER (II) ION, GLYCEROL, ... | | Authors: | Hagelueken, G. | | Deposit date: | 2013-07-04 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | High-Resolution Crystal Structure of Spin Labelled (T21R1) Azurin from Pseudomonas Aeruginosa: A Challenging Structural Benchmark for in Silico Spin Labelling Algorithms.
Bmc Struct.Biol., 14, 2014
|
|
6HVH
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 1 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-(oxan-4-yl)pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Jakubiec, K, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and Structure-Activity Relationships of N-Aryl 6-Aminoquinoxalines as Potent PFKFB3 Kinase Inhibitors.
ChemMedChem, 14, 2019
|
|
4NL9
 
 | |
