7WK5
 
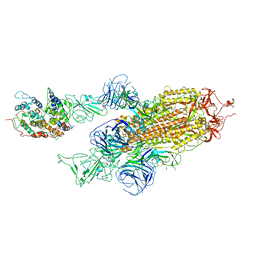 | | Cryo-EM structure of Omicron S-ACE2, C2 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Han, W.Y, Wang, Y.F. | | Deposit date: | 2022-01-08 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization
Biorxiv, 2022
|
|
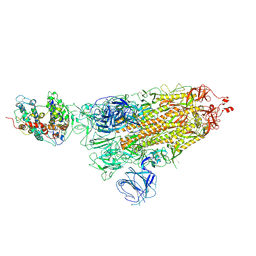
7WK4
 
 | |
7WK3
 
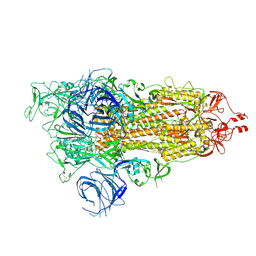 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
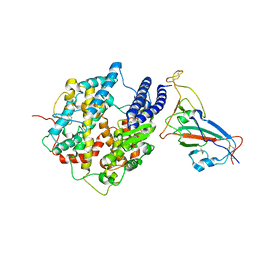
7WK6
 
 | |
5UKP
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.1 Fab | | Descriptor: | DH522.1 Fab fragment heavy chain, DH522.1 Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
5UKN
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522UCA Fab | | Descriptor: | CHLORIDE ION, DH522UCA Fab fragment heavy chain, DH522UCA Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
6MAR
 
 | |
7WM0
 
 | |
7XCK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Xie, Y.F, Liu, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (two-RBD-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | Authors: | Zhao, Z, Qi, J, Gao, F.G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with human ACE2 ectodomain (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7WT9
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab 9A8 | | Descriptor: | Heavy chain of Fab 9A8, Light chain of Fab 9A8, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT7
 
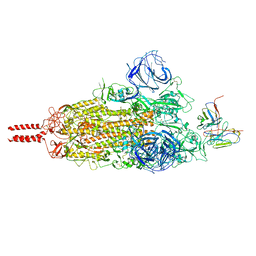 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT8
 
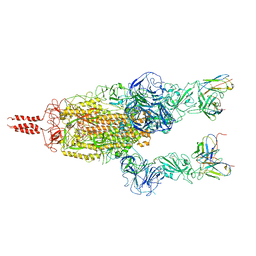 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
5UKR
 
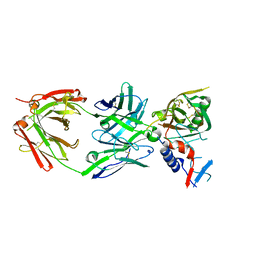 | |
5UKO
 
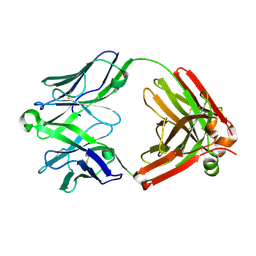 | | Structure of unliganded anti-gp120 CD4bs antibody DH522IA Fab | | Descriptor: | DH522IA Fab fragment heavy chain, DH522IA Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
7XCA
 
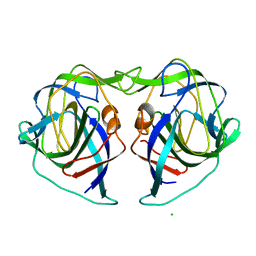 | | Crystal structure of the porcine astrovirus capsid spike domain | | Descriptor: | CHLORIDE ION, Capsid protein spike domain, MAGNESIUM ION | | Authors: | Pan, L.X, Zhang, W.C, Yang, D.F, Yang, L.Y, Liu, H. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure analysis of porcine astrovirus capsid spike and spike/neutralizing antibody complexes.
To Be Published
|
|
7Y7J
 
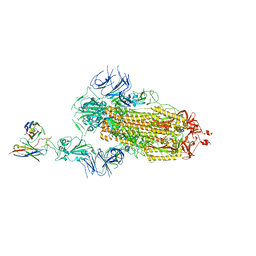 | | SARS-CoV-2 S trimer in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7Y7K
 
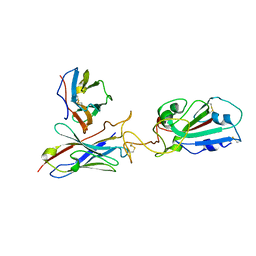 | | SARS-CoV-2 RBD in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, Spike protein S1 | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
5UKQ
 
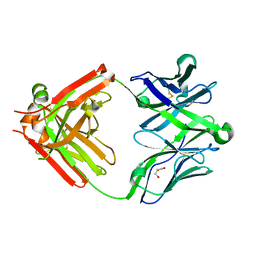 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.2 Fab | | Descriptor: | DH522.2 Fab fragment heavy chain, DH522.2 Fab fragment light chain, GLYCEROL | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
8VGW
 
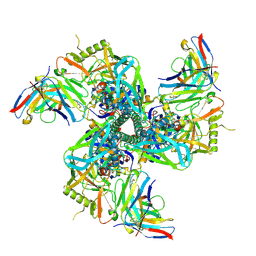 | | VRC01 Fab bound to the HIV-1 CH848 DE3 SOSIP | | Descriptor: | CH848 DE3 SOSIP gp120, CH848 DE3 SOSIP gp41, VRC01 Fab Heavy Chain, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-29 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
8VH1
 
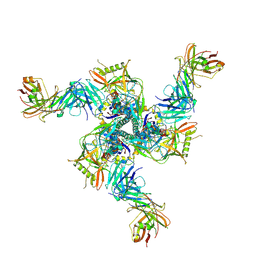 | | CH235.12 Fab bound to the HIV-1 CH505.M5 SOSIP | | Descriptor: | CH235.I60 Fab Heavy Chain, CH235.I60 Fab Light Chain, CH505.M5 SOSIP gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
8VH2
 
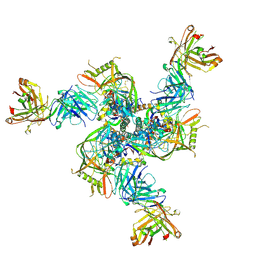 | | CH235.12 Fab bound to the HIV-1 CH505.M5 SOSIP | | Descriptor: | CH235.12 Fab Heavy Chain, CH235.12 Fab Light Chain, CH505.M5.G458Y SOSIP gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
