9G5Y
 
 | |
9G62
 
 | |
1ETP
 
 | |
1EAP
 
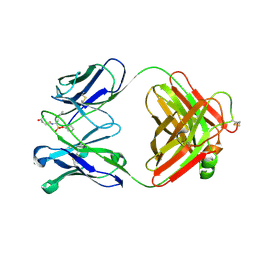 | | CRYSTAL STRUCTURE OF A CATALYTIC ANTIBODY WITH A SERINE PROTEASE ACTIVE SITE | | Descriptor: | IGG2B-KAPPA 17E8 FAB (HEAVY CHAIN), IGG2B-KAPPA 17E8 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE | | Authors: | Zhou, G.W, Guo, J, Huang, W, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1994-08-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a catalytic antibody with a serine protease active site.
Science, 265, 1994
|
|
5LHL
 
 | |
5LIG
 
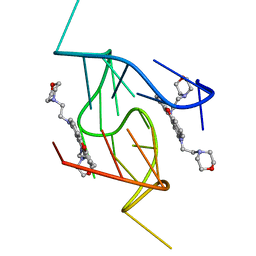 | | G-Quadruplex formed at the 5'-end of NHEIII_1 Element in human c-MYC promoter bound to triangulenium based fluorescence probe DAOTA-M2 | | Descriptor: | 8,12-bis(2-morpholinoethyl)-8H-benzo[ij]xantheno[1,9,8-cdef][2,7]naphthyridin-12-iumhexafluorophosphate, DNA (5'-D(*TP*AP*GP*GP*GP*AP*GP*GP*GP*TP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3') | | Authors: | Kotar, A, Wang, B, Shivalingam, A, Gonzalez-Garcia, J, Vilar, R, Plavec, J. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Triangulenium-Based Long-Lived Fluorescence Probe Bound to a G-Quadruplex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1E5S
 
 | | Proline 3-hydroxylase (type II) - Iron form | | Descriptor: | FE (II) ION, PROLINE OXIDASE, SULFATE ION | | Authors: | Clifton, I.J, Hsueh, L.C, Baldwin, J.E, Schofield, C.J, Harlos, K. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent oxygenases.
Eur.J.Biochem., 268, 2001
|
|
5LIO
 
 | |
5LWR
 
 | | Endothiapepsin in complex with a derivative of fragment 177 | | Descriptor: | 1,2-ETHANEDIOL, 4-[12-[(1-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazin-3-ium-3-yl)methyl]-10,11-dimethyl-3,4,6,7,11-pentazatricyclo[7.3.0.0^{2,6}]dodeca-1(12),2,4,7,9-pentaen-5-yl]-1,2,5-trimethyl-pyrrole-3-carbaldehyde, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1EM8
 
 | | Crystal structure of chi and psi subunit heterodimer from DNA POL III | | Descriptor: | DNA POLYMERASE III CHI SUBUNIT, DNA POLYMERASE III PSI SUBUNIT | | Authors: | Gulbis, J.M, Finkelstein, J, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2000-03-16 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex.
Eur.J.Biochem., 271, 2004
|
|
1ENZ
 
 | | CRYSTAL STRUCTURE AND FUNCTION OF THE ISONIAZID TARGET OF MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dessen, A, Quemard, A, Blanchard, J.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1995-01-27 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis.
Science, 267, 1995
|
|
9G99
 
 | |
5LXC
 
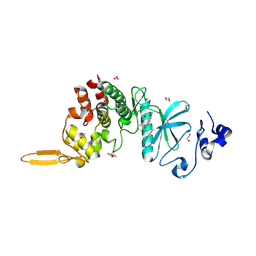 | | Crystal structure of DYRK2 in complex with EHT 5372 (Compound 1) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2,4-dichlorophenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
1EOB
 
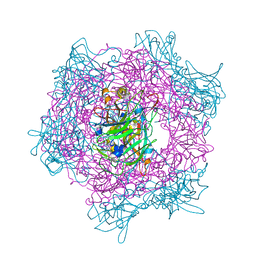 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE IN COMPLEX WITH 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
5LIW
 
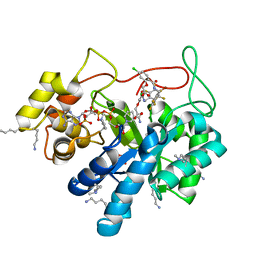 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK319 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
9G57
 
 | |
1E6M
 
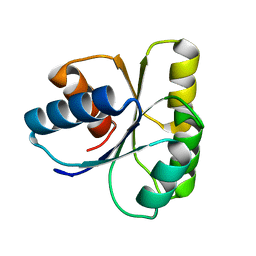 | | TWO-COMPONENT SIGNAL TRANSDUCTION SYSTEM D57A MUTANT OF CHEY | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
1E6U
 
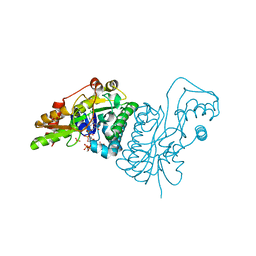 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-08-23 | | Release date: | 2000-10-22 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
9G6M
 
 | | Xylose Isomerase collected at 30C using time-resolved serial synchrotron crystallography with Glucose at 60 seconds | | Descriptor: | D-glucose, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Schulz, E.C, Prester, A, Stetten, D.V, Gore, G, Hatton, C.E, Bartels, K, Leimkohl, J.P, Schikora, H, Ginn, H.M, Tellkamp, F, Mehrabi, P. | | Deposit date: | 2024-07-18 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the modulation of enzyme kinetics by multi-temperature, time-resolved serial crystallography.
Nat Commun, 16, 2025
|
|
5LY8
 
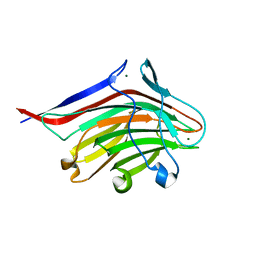 | | Structure of the CBM2 module of Lactobacillus casei BL23 phage J-1 evolved Dit. | | Descriptor: | MAGNESIUM ION, Tail component | | Authors: | Cambillau, C, Spinelli, S, Dieterle, M.-E, Piuri, M. | | Deposit date: | 2016-09-26 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Evolved distal tail carbohydrate binding modules of Lactobacillus phage J-1: a novel type of anti-receptor widespread among lactic acid bacteria phages.
Mol. Microbiol., 104, 2017
|
|
1E7K
 
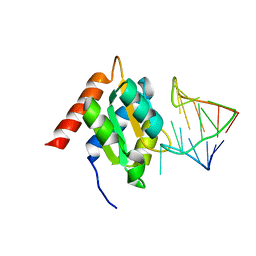 | | Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment | | Descriptor: | 15.5 KD RNA BINDING PROTEIN, RNA (5'-R(*GP*CP*CP*AP*AP*UP*GP*AP*GP*GP*UP*UP*UP* AP*UP*CP*CP*GP*AP*GP*G*C(-3') | | Authors: | Vidovic, I, Nottrott, S, Harthmuth, K, Luhrmann, R, Ficner, R. | | Deposit date: | 2000-08-29 | | Release date: | 2001-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Spliceosomal 15.5Kd Protein Bound to a U4 Snrna Fragment
Mol.Cell, 6, 2000
|
|
1EOJ
 
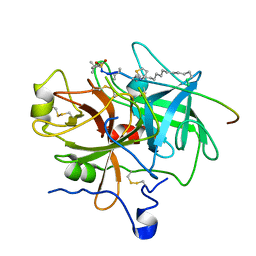 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P798 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
9G6P
 
 | | Xylose Isomerase collected at 50C using time-resolved serial synchrotron crystallography with Glucose at 60 seconds | | Descriptor: | D-glucose, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Schulz, E.C, Prester, A, Stetten, D.V, Gore, G, Hatton, C.E, Bartels, K, Leimkohl, J.P, Schikora, H, Ginn, H.M, Tellkamp, F, Mehrabi, P. | | Deposit date: | 2024-07-18 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the modulation of enzyme kinetics by multi-temperature, time-resolved serial crystallography.
Nat Commun, 16, 2025
|
|
1EP2
 
 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B COMPLEXED WITH OROTATE | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
