1EFL
 
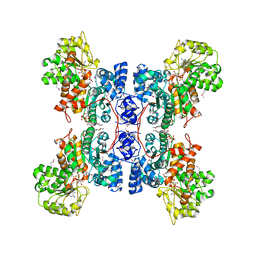 | | HUMAN MALIC ENZYME IN A QUATERNARY COMPLEX WITH NAD, MG, AND TARTRONATE | | Descriptor: | MAGNESIUM ION, MALIC ENZYME, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
5LJL
 
 | | Streptococcus pneumonia TIGR4 flavodoxin: structural and biophysical characterization of a novel drug target | | Descriptor: | Flavodoxin, PHOSPHATE ION | | Authors: | Rodriguez-Cardenas, A, Rojas, A.L, Velazquez-Campoy, A, Hurtado-Guerrero, R, Sancho, J. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Streptococcus pneumoniae TIGR4 Flavodoxin: Structural and Biophysical Characterization of a Novel Drug Target.
Plos One, 11, 2016
|
|
5LYD
 
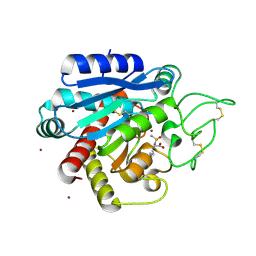 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-6-azanyl-2-(sulfamoylamino)hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-27 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
1EIA
 
 | |
5LYL
 
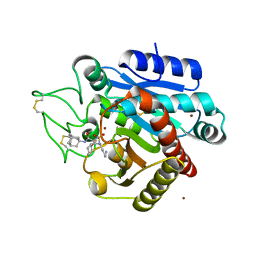 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-2-[[(2~{S})-1-(1-adamantylamino)-3-cyclohexyl-1-oxidanylidene-propan-2-yl]sulfamoylamino]-6-azanyl-hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
1EII
 
 | | NMR STRUCTURE OF HOLO CELLULAR RETINOL-BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II, RETINOL | | Authors: | Lu, J, Lin, C.L, Tang, C, Ponder, J.W, Kao, J.L, Cistola, D.P, Li, E. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding of retinol induces changes in rat cellular retinol-binding protein II conformation and backbone dynamics.
J.Mol.Biol., 300, 2000
|
|
1EKO
 
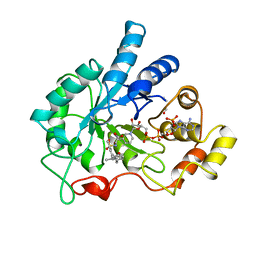 | | PIG ALDOSE REDUCTASE COMPLEXED WITH IDD384 INHIBITOR | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,6-DIMETHYL-4-(2-O-TOLYL-ACETYLAMINO)-BENZENESULFONYL]-GLYCINE | | Authors: | Podjarny, A. | | Deposit date: | 2000-03-09 | | Release date: | 2000-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of human aldose reductase bound to the inhibitor IDD384.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EKW
 
 | | NMR STRUCTURE OF A DNA THREE-WAY JUNCTION | | Descriptor: | DNA (5'-D(*CP*GP*GP*TP*GP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*TP*CP*GP*CP*AP*GP*C)-3') | | Authors: | Thiviyanathan, V, Luxon, B.A, Leontis, N.B, Donne, D, Gorenstein, D.G. | | Deposit date: | 2000-03-09 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hybrid-hybrid matrix structural refinement of a DNA three-way junction from 3D NOESY-NOESY.
J.Biomol.NMR, 14, 1999
|
|
7S49
 
 | | Crystal Structure of Inhibitor-bound Galactokinase | | Descriptor: | (4R)-2-[(1,3-benzoxazol-2-yl)amino]-4-(4-chloro-1H-pyrazol-5-yl)-4,6,7,8-tetrahydroquinazolin-5(1H)-one, Galactokinase, PHOSPHATE ION, ... | | Authors: | Whitby, F.G. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
1EL3
 
 | | HUMAN ALDOSE REDUCTASE COMPLEXED WITH IDD384 INHIBITOR | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,6-DIMETHYL-4-(2-O-TOLYL-ACETYLAMINO)-BENZENESULFONYL]-GLYCINE | | Authors: | Podjarny, A. | | Deposit date: | 2000-03-13 | | Release date: | 2000-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of human aldose reductase bound to the inhibitor IDD384.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5LK6
 
 | |
1ELQ
 
 | | CRYSTAL STRUCTURE OF THE CYSTINE C-S LYASE C-DES | | Descriptor: | L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5LKT
 
 | | Crystal structure of the p300 acetyltransferase catalytic core with butyryl-coenzyme A. | | Descriptor: | Butyryl Coenzyme A, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-24 | | Release date: | 2016-11-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5LZ5
 
 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 2-fluoranyl-5-[2-[(4~{S})-4-methyl-2-oxidanylidene-4-phenyl-pyrrolidin-1-yl]ethoxy]benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A.J.A, Day, P.J. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
1EMG
 
 | | GREEN FLUORESCENT PROTEIN (65-67 REPLACED BY CRO, S65T SUBSTITUTION, Q80R) | | Descriptor: | PROTEIN (GREEN FLUORESCENT PROTEIN) | | Authors: | Elsliger, M.A, Wachter, R.M, Kallio, K, Hanson, G.T, Remington, S.J. | | Deposit date: | 1998-11-12 | | Release date: | 1999-05-12 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and spectral response of green fluorescent protein variants to changes in pH.
Biochemistry, 38, 1999
|
|
5LZL
 
 | | Pyrobaculum calidifontis 5-aminolaevulinic acid dehydratase | | Descriptor: | Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Azim, N, Erskine, P.T, Guo, J, Cooper, J.B. | | Deposit date: | 2016-09-30 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1EMT
 
 | | FAB ANTIBODY FRAGMENT OF AN C60 ANTIFULLERENE ANTIBODY | | Descriptor: | IGG ANTIBODY (HEAVY CHAIN), IGG ANTIBODY (LIGHT CHAIN) | | Authors: | Braden, B.C, Goldbaum, F.A, Chen, B.-X, Erlanger, B.F, Kirschner, A.N, Wilson, S.R. | | Deposit date: | 2000-03-17 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystal structure of an anti-Buckminsterfullerene antibody fab fragment: biomolecular recognition of C(60).
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ENC
 
 | | CRYSTAL STRUCTURES OF THE BINARY CA2+ AND PDTP COMPLEXES AND THE TERNARY COMPLEX OF THE ASP 21->GLU MUTANT OF STAPHYLOCOCCAL NUCLEASE. IMPLICATIONS FOR CATALYSIS AND LIGAND BINDING | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Libson, A, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-02-14 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the binary Ca2+ and pdTp complexes and the ternary complex of the Asp21-->Glu mutant of staphylococcal nuclease. Implications for catalysis and ligand binding.
Biochemistry, 33, 1994
|
|
5LKZ
 
 | | Crystal structure of the p300 acetyltransferase catalytic core with crotonyl-coenzyme A. | | Descriptor: | CROTONYL COENZYME A, GLYCEROL, Histone acetyltransferase p300,Histone acetyltransferase p300, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
2PC8
 
 | | E292Q mutant of EXO-B-(1,3)-Glucanase from Candida Albicans in complex with two separately bound glucopyranoside units at 1.8 A | | Descriptor: | Hypothetical protein XOG1, beta-D-glucopyranose | | Authors: | Cutfield, S.M, Cutfield, J.F, Patrick, W.M. | | Deposit date: | 2007-03-29 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Carbohydrate binding sites in Candida albicans exo-beta-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance.
Febs J., 277, 2010
|
|
5LLJ
 
 | |
1ENN
 
 | | SOLVENT ORGANIZATION IN AN OLIGONUCLEOTIDE CRYSTAL: THE STRUCTURE OF D(GCGAATTCG)2 AT ATOMIC RESOLUTION | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Soler-Lopez, M, Malinina, L, Subirana, J.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Solvent organization in an oligonucleotide crystal. The structure of d(GCGAATTCG)2 at atomic resolution.
J.Biol.Chem., 275, 2000
|
|
1EO0
 
 | | CONSERVED DOMAIN COMMON TO TRANSCRIPTION FACTORS TFIIS, ELONGIN A, CRSP70 | | Descriptor: | TRANSCRIPTION ELONGATION FACTOR S-II | | Authors: | Booth, V, Koth, C, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70
J.Biol.Chem., 275, 2000
|
|
9G60
 
 | |
1EOK
 
 | | CRYSTAL STRUCTURE OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3 | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3, SULFATE ION | | Authors: | Waddling, C.A, Plummer Jr, T.H, Tarentino, A.L, Van Roey, P. | | Deposit date: | 2000-03-23 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3).
Biochemistry, 39, 2000
|
|
