1ETY
 
 | |
1ETZ
 
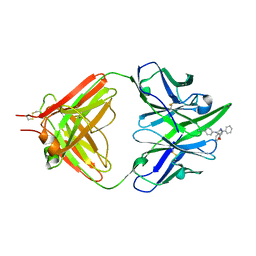 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
1EU1
 
 | | THE CRYSTAL STRUCTURE OF RHODOBACTER SPHAEROIDES DIMETHYLSULFOXIDE REDUCTASE REVEALS TWO DISTINCT MOLYBDENUM COORDINATION ENVIRONMENTS. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Li, H.K, Temple, K, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2000-04-13 | | Release date: | 2000-08-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A Crystal Structure of Rhodobacter sphaeroides Dimethylsulfoxide Reductase Reveals Two Distinct Molybdenum Coordination Environments
J.Am.Chem.Soc., 122, 2000
|
|
1EU2
 
 | |
1EU3
 
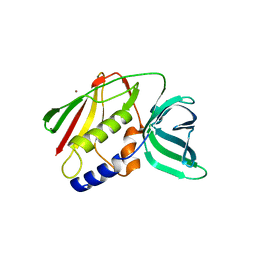 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SMEZ-2 (ZINC BOUND) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, SUPERANTIGEN SMEZ-2, ... | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-13 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1EU4
 
 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SPE-H (ZINC BOUND) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | SUPERANTIGEN SPE-H, ZINC ION | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-13 | | Release date: | 2000-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1EU5
 
 | | STRUCTURE OF E. COLI DUTPASE AT 1.45 A | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, GLYCEROL | | Authors: | Gonzalez, A, Larsson, G, Persson, R, Cedergren-Zeppezauer, E. | | Deposit date: | 2000-04-13 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic resolution structure of Escherichia coli dUTPase determined ab initio.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EU6
 
 | |
1EU8
 
 | | STRUCTURE OF TREHALOSE MALTOSE BINDING PROTEIN FROM THERMOCOCCUS LITORALIS | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, TREHALOSE/MALTOSE BINDING PROTEIN, ... | | Authors: | Diez, J, Diederichs, K, Greller, G, Horlacher, R, Boos, W, Welte, W. | | Deposit date: | 2000-04-14 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a liganded trehalose/maltose-binding protein from the hyperthermophilic Archaeon Thermococcus litoralis at 1.85 A.
J.Mol.Biol., 305, 2001
|
|
1EUA
 
 | | SCHIFF BASE INTERMEDIATE IN KDPG ALDOLASE FROM ESCHERICHIA COLI | | Descriptor: | ACETATE ION, KDPG ALDOLASE, PYRUVIC ACID, ... | | Authors: | Allard, J, Grochulski, P, Sygusch, J. | | Deposit date: | 2000-04-14 | | Release date: | 2001-02-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Covalent intermediate trapped in 2-keto-3-deoxy-6- phosphogluconate (KDPG) aldolase structure at 1.95-A resolution.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1EUB
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED TO A POTENT NON-PEPTIDIC SULFONAMIDE INHIBITOR | | Descriptor: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | Authors: | Zhang, X, Gonnella, N.C, Koehn, J, Pathak, N, Ganu, V, Melton, R, Parker, D, Hu, S.I, Nam, K.Y. | | Deposit date: | 2000-04-14 | | Release date: | 2001-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human collagenase-3 (MMP-13) complexed to a potent non-peptidic sulfonamide inhibitor: binding comparison with stromelysin-1 and collagenase-1.
J.Mol.Biol., 301, 2000
|
|
1EUC
 
 | | CRYSTAL STRUCTURE OF DEPHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE | | Descriptor: | PHOSPHATE ION, SUCCINYL-COA SYNTHETASE, ALPHA CHAIN, ... | | Authors: | Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 2000-04-14 | | Release date: | 2000-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA synthetase.
J.Mol.Biol., 299, 2000
|
|
1EUD
 
 | | CRYSTAL STRUCTURE OF PHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE | | Descriptor: | SUCCINYL-COA SYNTHETASE, ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 2000-04-14 | | Release date: | 2000-07-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA synthetase.
J.Mol.Biol., 299, 2000
|
|
1EUE
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oganesyan, V, Zhang, X. | | Deposit date: | 2000-04-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of redox potential in electron transfer proteins: effects of complex formation on the active site microenvironment of cytochrome b5.
FARADAY DISC.CHEM.SOC, 116, 2001
|
|
1EUF
 
 | | BOVINE DUODENASE(NEW SERINE PROTEASE), CRYSTAL STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DUODENASE, PHOSPHATE ION | | Authors: | Pletnev, V.Z, Zamolodchikova, T.S, Pangborn, W.A, Duax, W.L. | | Deposit date: | 2000-04-14 | | Release date: | 2001-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of bovine duodenase, a serine protease, with dual trypsin and chymotrypsin-like specificities.
Proteins, 41, 2000
|
|
1EUG
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | Descriptor: | PROTEIN (GLYCOSYLASE) | | Authors: | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
1EUH
 
 | | APO FORM OF A NADP DEPENDENT ALDEHYDE DEHYDROGENASE FROM STREPTOCOCCUS MUTANS | | Descriptor: | NADP DEPENDENT NON PHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1998-11-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 290, 1999
|
|
1EUI
 
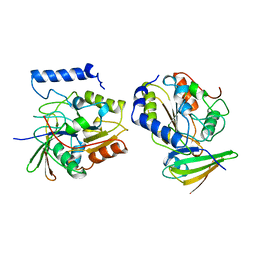 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Authors: | Ravishankar, R, Sagar, M.B, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray analysis of a complex of Escherichia coli uracil DNA glycosylase (EcUDG) with a proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG.
Nucleic Acids Res., 26, 1998
|
|
1EUJ
 
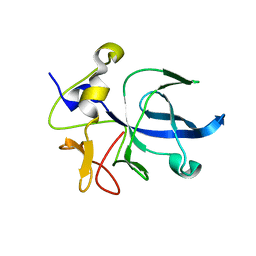 | | A NOVEL ANTI-TUMOR CYTOKINE CONTAINS A RNA-BINDING MOTIF PRESENT IN AMINOACYL-TRNA SYNTHETASES | | Descriptor: | ENDOTHELIAL MONOCYTE ACTIVATING POLYPEPTIDE 2 | | Authors: | Kim, Y, Shin, J, Li, R, Cheong, C, Kim, S. | | Deposit date: | 2000-04-17 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel anti-tumor cytokine contains an RNA binding motif present in aminoacyl-tRNA synthetases.
J.Biol.Chem., 275, 2000
|
|
1EUM
 
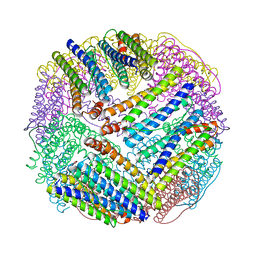 | | CRYSTAL STRUCTURE OF THE E.COLI FERRITIN ECFTNA | | Descriptor: | FERRITIN 1 | | Authors: | Stillman, T.J, Hempstead, P.D, Artymiuk, P.J, Andrews, S.C, Hudson, A.J, Treffry, A, Guest, J.R, Harrison, P.M. | | Deposit date: | 2000-04-17 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The high-resolution X-ray crystallographic structure of the ferritin (EcFtnA) of Escherichia coli; comparison with human H ferritin (HuHF) and the structures of the Fe(3+) and Zn(2+) derivatives.
J.Mol.Biol., 307, 2001
|
|
1EUN
 
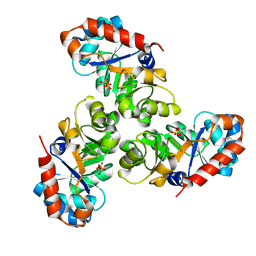 | |
1EUO
 
 | | Crystal structure of nitrophorin 2 (prolixin-S) | | Descriptor: | AMMONIA, NITROPHORIN 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Andersen, J.F, Montfort, W.R. | | Deposit date: | 2000-04-17 | | Release date: | 2000-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of nitrophorin 2. A trifunctional antihemostatic protein from the saliva of Rhodnius prolixus
J.Biol.Chem., 275, 2000
|
|
1EUP
 
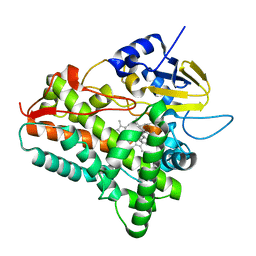 | |
1EUQ
 
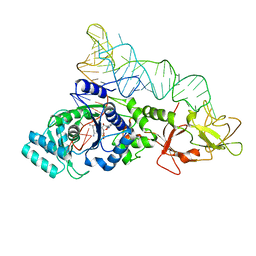 | | CRYSTAL STRUCTURE OF GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH A TRNA-GLN MUTANT AND AN ACTIVE-SITE INHIBITOR | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, GLUTAMINYL TRNA, GLUTAMINYL-TRNA SYNTHETASE | | Authors: | Sherlin, L.D, Bullock, T.L, Newberry, K.J, Lipman, R.S.A, Hou, Y.-M, Beijer, B, Sproat, B.S, Perona, J.J. | | Deposit date: | 2000-04-17 | | Release date: | 2000-06-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases.
J.Mol.Biol., 299, 2000
|
|
1EUR
 
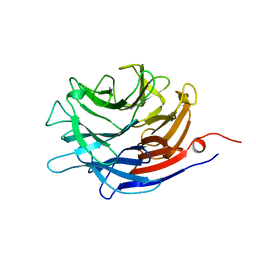 | | SIALIDASE | | Descriptor: | SIALIDASE | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
