5LUJ
 
 | |
1E5V
 
 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
5LDV
 
 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | CALCIUM ION, MOMP porin, N-OCTANE, ... | | Authors: | Wallat, G.D, Ferrara, L.M.G, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
1EHS
 
 | | THE STRUCTURE OF ESCHERICHIA COLI HEAT-STABLE ENTEROTOXIN B BY NUCLEAR MAGNETIC RESONANCE AND CIRCULAR DICHROISM | | Descriptor: | HEAT-STABLE ENTEROTOXIN B | | Authors: | Sukumar, M, Rizo, J, Wall, M, Dreyfus, L.A, Kupersztoch, Y.M, Gierasch, L.M. | | Deposit date: | 1995-06-13 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structure of Escherichia coli heat-stable enterotoxin b by nuclear magnetic resonance and circular dichroism.
Protein Sci., 4, 1995
|
|
1EGO
 
 | | NMR STRUCTURE OF OXIDIZED ESCHERICHIA COLI GLUTAREDOXIN: COMPARISON WITH REDUCED E. COLI GLUTAREDOXIN AND FUNCTIONALLY RELATED PROTEINS | | Descriptor: | GLUTAREDOXIN | | Authors: | Xia, T.-H, Bushweller, J.H, Sodano, P, Billeter, M, Bjornberg, O, Holmgren, A, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
1ERA
 
 | |
5LVF
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Thr4 phosphorylated CTD peptide | | Descriptor: | PRO-SER-TYR-SER-PRO-PTH-SER-PRO-SER-TYR-SER-PRO-THR-SER-PRO-SER, Regulator of Ty1 transposition protein 103 | | Authors: | Jasnovidova, O, Kubicek, K, Krejcikova, M, Stefl, R. | | Deposit date: | 2016-09-14 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural insight into recognition of phosphorylated threonine-4 of RNA polymerase II C-terminal domain by Rtt103p.
EMBO Rep., 18, 2017
|
|
5LWE
 
 | | Crystal structure of the human CC chemokine receptor type 9 (CCR9) in complex with vercirnon | | Descriptor: | C-C chemokine receptor type 9, CHOLESTEROL, MALONATE ION, ... | | Authors: | Oswald, C, Rappas, M, Kean, J, Dore, A.S, Errey, J.C, Bennett, K, Deflorian, F, Christopher, J.A, Jazayeri, A, Mason, J.S, Congreve, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intracellular allosteric antagonism of the CCR9 receptor.
Nature, 540, 2016
|
|
1E7L
 
 | | Endonuclease VII (EndoVII) N62D mutant from phage T4 | | Descriptor: | RECOMBINATION ENDONUCLEASE VII, SULFATE ION, ZINC ION | | Authors: | Raaijmakers, H.C.A, Vix, O, Toro, I, Suck, D. | | Deposit date: | 2000-08-29 | | Release date: | 2001-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Conformational Flexibility in T4 Endonuclease Vii Revealed by Crystallography: Implications for Substrate Binding and Cleavage
J.Mol.Biol., 308, 2001
|
|
5LWS
 
 | | Endothiapepsin in complex with fragment 177 and a derivative thereof | | Descriptor: | 4-[12-[(1-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazin-3-ium-3-yl)methyl]-10,11-dimethyl-3,4,6,7,11-pentazatricyclo[7.3.0.0^{2,6}]dodeca-1(12),2,4,7,9-pentaen-5-yl]-1,2,5-trimethyl-pyrrole-3-carbaldehyde, 4-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazine, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1E82
 
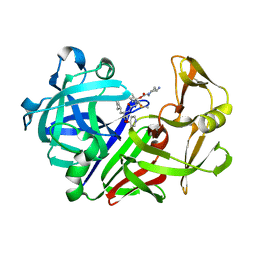 | | Endothiapepsin complex with renin inhibitor MERCK-KGAA-EMD59601 | | Descriptor: | (2S)-1-{[(2R)-1-{[(2S,3R)-1-cyclohexyl-3-hydroxy-4-(pyridin-4-yloxy)butan-2-yl]amino}-3-(methylsulfanyl)-1-oxopropan-2-yl]amino}-1-oxo-3-phenylpropan-2-yl 4-aminopiperidine-1-carboxylate, ENDOTHIAPEPSIN | | Authors: | Read, J.A, Cooper, J.B, Toldo, L, Rippmann, F, Raddatz, P. | | Deposit date: | 2000-09-15 | | Release date: | 2000-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refinement of Four Endothiapepsin Inhibitor Complexes. Crystallographic Studies of Cytochrome Ch from Methylobacterium Extorquens and Inhibitor Complexes of Aspartic Proteinases.
Thesis, 1999
|
|
5LE6
 
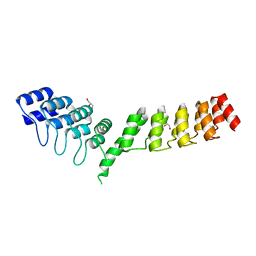 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_09_D12 | | Descriptor: | DD_D12_09_D12, GLYCEROL, SULFATE ION | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
1EC1
 
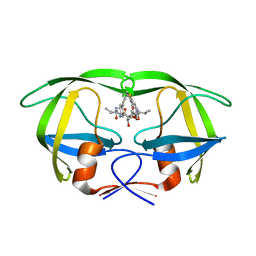 | | HIV-1 protease in complex with the inhibitor BEA409 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DI-4-THIOPHEN-3-YL-BENZYL]-GLUCARYL]-DI-[VALYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
5LX3
 
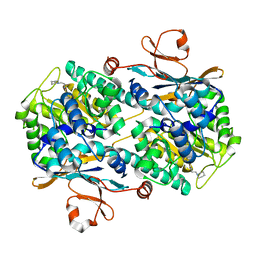 | | CRYSTAL STRUCTURE OF VISFATIN IN COMPLEX WITH SAR154782. | | Descriptor: | 6-[4-[(6-azanylpyridin-3-yl)methylcarbamoylamino]-3-fluoranyl-phenyl]-2-(ethylamino)-~{N}-(2-piperidin-1-ylethyl)pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase | | Authors: | Bertrand, T, Marquette, J.P. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF VISFATIN IN COMPLEX WITH SAR154782.
To Be Published
|
|
1ECF
 
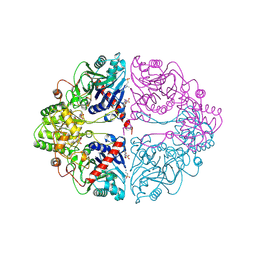 | |
5LXI
 
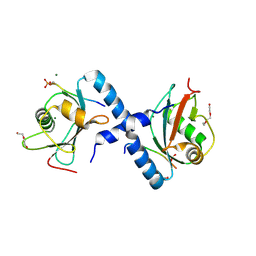 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | 1,2-ETHANEDIOL, Cysteine protease ATG4B, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
1ED3
 
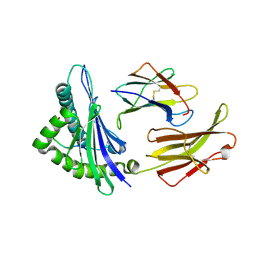 | | CRYSTAL STRUCTURE OF RAT MINOR HISTOCOMPATIBILITY ANTIGEN COMPLEX RT1-AA/MTF-E. | | Descriptor: | BETA-2-MICROGLOBULIN, CLASS I MAJOR HISTOCOMPATIBILITY ANTIGEN RT1-AA, PEPTIDE MTF-E (13N3E) | | Authors: | Speir, J.A, Stevens, J, Joly, E, Butcher, G.W, Wilson, I.A. | | Deposit date: | 2000-01-26 | | Release date: | 2001-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa.
Immunity, 14, 2001
|
|
5LGN
 
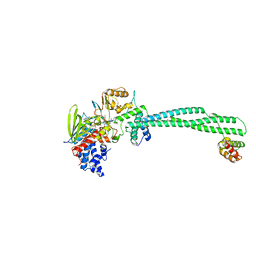 | | Thieno[3,2-b]pyrrole-5-carboxamides as Novel Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1: Compound 19 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Mattevi, A, Ciossani, G. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 1: High-Throughput Screening and Preliminary Exploration.
J. Med. Chem., 60, 2017
|
|
1EDO
 
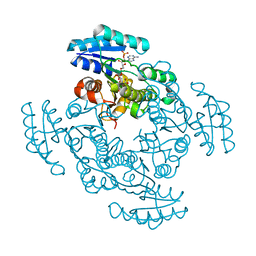 | | THE X-RAY STRUCTURE OF BETA-KETO ACYL CARRIER PROTEIN REDUCTASE FROM BRASSICA NAPUS COMPLEXED WITH NADP+ | | Descriptor: | BETA-KETO ACYL CARRIER PROTEIN REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Fisher, M, Kroon, J.T, Martindale, W, Stuitje, A.R, Slabas, A.R, Rafferty, J.B. | | Deposit date: | 2000-01-28 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray structure of Brassica napus beta-keto acyl carrier protein reductase and its implications for substrate binding and catalysis.
Structure Fold.Des., 8, 2000
|
|
1EE1
 
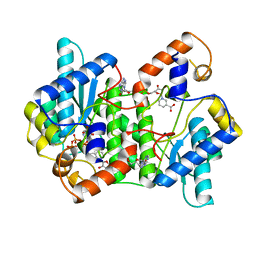 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE ATP, TWO MOLECULES DEAMIDO-NAD+ AND ONE MG2+ ION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, Delucas, L. | | Deposit date: | 2000-01-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5LGX
 
 | |
1EEN
 
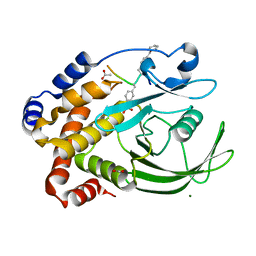 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-D-A-D-BPA-PTYR-L-I-P-Q-Q-G | | Descriptor: | ACETIC ACID, ALA-ASP-PBF-PTR-LEU-ILE-PRO, MAGNESIUM ION, ... | | Authors: | Puius, Y.A, Zhao, Y, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
5LXR
 
 | | Structure of the minimal RBM7 - ZCCHC8 Complex | | Descriptor: | BROMIDE ION, CHLORIDE ION, RNA-binding protein 7, ... | | Authors: | Falk, S, Finogenova, K, Benda, C, Conti, E. | | Deposit date: | 2016-09-22 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the RBM7-ZCCHC8 core of the NEXT complex reveals connections to splicing factors.
Nat Commun, 7, 2016
|
|
1EF9
 
 | | THE CRYSTAL STRUCTURE OF METHYLMALONYL COA DECARBOXYLASE COMPLEXED WITH 2S-CARBOXYPROPYL COA | | Descriptor: | 2-CARBOXYPROPYL-COENZYME A, METHYLMALONYL COA DECARBOXYLASE | | Authors: | Benning, M.M, Haller, T, Gerlt, J.A, Holden, H.M. | | Deposit date: | 2000-02-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli.
Biochemistry, 39, 2000
|
|
