4WWK
 
 | | Crystal structure of human TCR Alpha Chain-TRAV12-3, Beta Chain-TRBV6-5, Antigen-presenting molecule CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, A, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
1LZR
 
 | |
3QTR
 
 | | CDK2 in complex with inhibitor RC-1-148 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(phenylamino)-1,3-thiazol-5-yl](phenyl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3IAQ
 
 | | E. coli (lacz) beta-galactosidase (E416V) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Lo, S, Dugdale, M.L, Jeerh, N, Ku, T, Roth, N.J, Huber, R.E. | | Deposit date: | 2009-07-14 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Studies of Glu-416 variants of beta-galactosidase (E. coli) show that the active site Mg(2+) is not important for structure and indicate that the main role of Mg (2+) is to mediate optimization of active site chemistry
Protein J., 29, 2010
|
|
4R3Z
 
 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6LKB
 
 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
5O3D
 
 | | Human Brd2(BD2) mutant in complex with 9-ET | | Descriptor: | (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, CHLORIDE ION, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
4X7Q
 
 | | PIM2 kinase in complex with Compound 1s | | Descriptor: | 2-(2,6-difluorophenyl)-N-{4-[(3S)-pyrrolidin-3-yloxy]pyridin-3-yl}-1,3-thiazole-4-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-2 | | Authors: | Marcotte, D.J, Silvian, L.F. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-based design of low-nanomolar PIM kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5CQH
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
6UF9
 
 | | S4 symmetric peptide design number 1, Tim apo form | | Descriptor: | S4-1, Tim apo-form, SULFATE ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
3QXQ
 
 | | Structure of the bacterial cellulose synthase subunit Z in complex with cellopentaose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Zimmer, J. | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Apo- and cellopentaose-bound structures of the bacterial cellulose synthase subunit BcsZ.
J.Biol.Chem., 286, 2011
|
|
2W70
 
 | |
7ESH
 
 | | Crystal structure of amylosucrase from Calidithermus timidus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase | | Authors: | Tian, Y, Hou, X, Ni, D, Xu, W, Guang, C, Zhang, W, Rao, Y, Mu, W. | | Deposit date: | 2021-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based interface engineering methodology in designing a thermostable amylose-forming transglucosylase
J.Biol.Chem., 298, 2022
|
|
3IKG
 
 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-(3-methylphenyl)propyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Parge, H, Ferre, R.A, Greasley, S, Matthews, D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3QU0
 
 | | CDK2 in complex with inhibitor RC-2-38 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(pyridin-3-ylcarbonyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
215L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | Descriptor: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
 | |
6AZN
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
3IRP
 
 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus at 1.50 angstrom resolution | | Descriptor: | GLYCEROL, POTASSIUM ION, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
9JQW
 
 | | The Crystal Structure of LCC6-Active from Biortus | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Leaf-branch compost cutinase, ... | | Authors: | Wan, T, Wang, F, Lv, Z, Lin, D, Pan, W, Sang, C, Sun, J. | | Deposit date: | 2024-09-28 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of LCC6-Active from Biortus
To Be Published
|
|
7EUT
 
 | | Crystal structures of 2-oxoglutarate dependent dioxygenase (CTB9) in complex with N-oxalylglycine | | Descriptor: | 1,2-ETHANEDIOL, 2-oxoglutarate (2-OG)-dependent dioxygenase, COPPER (II) ION, ... | | Authors: | Hou, X.D, Liu, X.Z, Yuan, Z.B, Yin, D.J, Rao, Y.J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Molecular Basis of the Unusual Seven-Membered Methylenedioxy Bridge Formation Catalyzed by Fe(II)/alpha-KG-Dependent Oxygenase CTB9
Acs Catalysis, 12, 2022
|
|
2AET
 
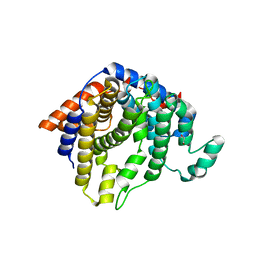 | | R304K trichodiene synthase: Complex with Mg, pyrophosphate, and (4S)-7-azabisabolene | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-07-23 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Role of arginine-304 in the diphosphate-triggered active site closure mechanism of trichodiene synthase.
Biochemistry, 44, 2005
|
|
7EUU
 
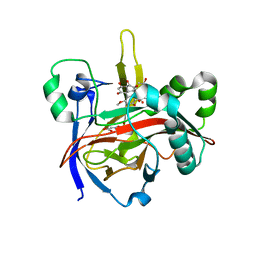 | | Crystal structures of 2-oxoglutarate dependent dioxygenase (CTB9) in complex with N-oxalylglycine and pre-cercosporin | | Descriptor: | 1,2-ETHANEDIOL, 2,6,11-trimethoxy-4,7,9-tris(oxidanyl)-1,12-bis[(2R)-2-oxidanylpropyl]perylene-3,10-dione, 2-oxoglutarate (2-OG)-dependent dioxygenase, ... | | Authors: | Hou, X.D, Liu, X.Z, Yuan, Z.B, Rao, Y.J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular Basis of the Unusual Seven-Membered Methylenedioxy Bridge Formation Catalyzed by Fe(II)/alpha-KG-Dependent Oxygenase CTB9
Acs Catalysis, 12, 2022
|
|
3ISM
 
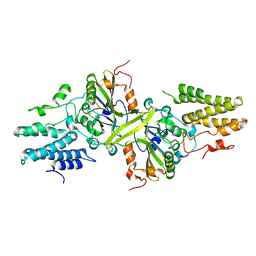 | | Crystal structure of the EndoG/EndoGI complex: Mechanism of EndoG inhibition | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CG4930, CG8862, ... | | Authors: | Loll, B, Gebhardt, M, Wahle, E, Meinhart, A. | | Deposit date: | 2009-08-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the EndoG/EndoGI complex: mechanism of EndoG inhibition.
Nucleic Acids Res., 37, 2009
|
|
