1EP2
 
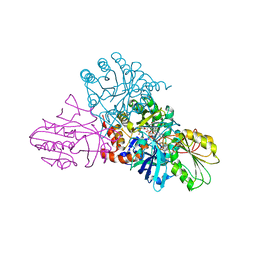 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B COMPLEXED WITH OROTATE | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
1EP3
 
 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B. DATA COLLECTED UNDER CRYOGENIC CONDITIONS. | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
1EP4
 
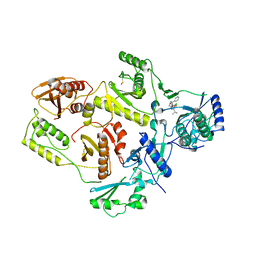 | | Crystal structure of HIV-1 reverse transcriptase in complex with S-1153 | | Descriptor: | 5-(3,5-DICHLOROPHENYL)THIO-4-ISOPROPYL-1-(PYRIDIN-4-YL-METHYL)-1H-IMIDAZOL-2-YL-METHYL CARBAMATE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Nichols, C, Bird, L.E, Fujiwara, T, Suginoto, H, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-03-27 | | Release date: | 2000-09-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding.
J.Biol.Chem., 275, 2000
|
|
1EP5
 
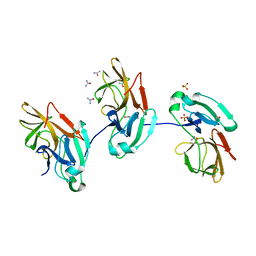 | |
1EP6
 
 | |
1EP7
 
 | | CRYSTAL STRUCTURE OF WT THIOREDOXIN H FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | THIOREDOXIN CH1, H-TYPE | | Authors: | Menchise, V, Corbier, C, Didierjean, C, Saviano, M, Benedetti, E, Jacquot, J.P, Aubry, A. | | Deposit date: | 2000-03-28 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism.
Biochem.J., 359, 2001
|
|
1EP8
 
 | | CRYSTAL STRUCTURE OF A MUTATED THIOREDOXIN, D30A, FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | THIOREDOXIN CH1, H-TYPE | | Authors: | Menchise, V, Corbier, C, Didierjean, C, Saviano, M, Benedetti, E, Jacquot, J.P, Aubry, A. | | Deposit date: | 2000-03-28 | | Release date: | 2001-12-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism.
Biochem.J., 359, 2001
|
|
1EP9
 
 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CONFORMATIONAL CHANGE | | Descriptor: | ORNITHINE TRANSCARBAMYLASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Morizono, H, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2000-03-28 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human ornithine transcarbamylase: crystallographic insights into substrate recognition and conformational changes.
Biochem.J., 354, 2001
|
|
1EPA
 
 | |
1EPB
 
 | |
1EPF
 
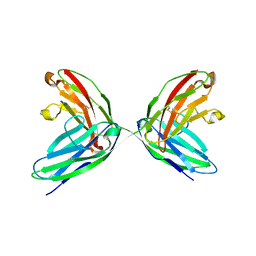 | | CRYSTAL STRUCTURE OF THE TWO N-TERMINAL IMMUNOGLOBULIN DOMAINS OF THE NEURAL CELL ADHESION MOLECULE (NCAM) | | Descriptor: | CALCIUM ION, PROTEIN (NEURAL CELL ADHESION MOLECULE) | | Authors: | Kasper, C, Rasmussen, H, Kastrup, J.S, Ikemizu, S, Jones, E.Y, Berezin, V, Bock, E, Larsen, I.K. | | Deposit date: | 2000-03-29 | | Release date: | 2000-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of cell-cell adhesion by NCAM.
Nat.Struct.Biol., 7, 2000
|
|
1EPG
 
 | |
1EPH
 
 | |
1EPI
 
 | |
1EPJ
 
 | |
1EPL
 
 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS1, PRO-LEU-GLU-PSA-ARG-LEU | | Authors: | Al-Karadaghi, S, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EPM
 
 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS2, THR-PHE-GLN-ALA-PSA-LEU-ARG-GLU, ... | | Authors: | Crawford, M, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EPN
 
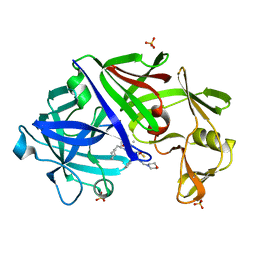 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R,2S)-1-(cyclohexylmethyl)-2-hydroxy-3-(1-methylethoxy)-3-oxopropyl]-S-methyl-L-cysteinamide, SULFATE ION | | Authors: | Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EPO
 
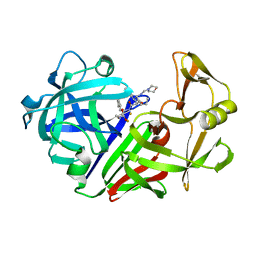 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH CP-81,282 (MOR PHE NLE CHF NME) | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide | | Authors: | Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct observation by X-ray analysis of the tetrahedral intermediate of aspartic proteinases.
Protein Sci., 1, 1992
|
|
1EPP
 
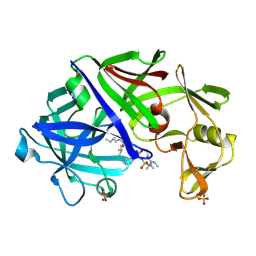 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-130,693 (MAS PHE LYS+MTF STA MBA) | | Descriptor: | ENDOTHIAPEPSIN, N-(dimethylsulfamoyl)-L-phenylalanyl-N-[(1S,2S)-2-hydroxy-4-{[(2S)-2-methylbutyl]amino}-1-(2-methylpropyl)-4-oxobutyl]-N~6~-(methylcarbamothioyl)-L-lysinamide, SULFATE ION | | Authors: | Wallace, B.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1EPQ
 
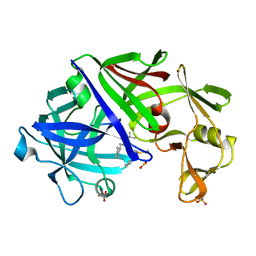 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-133,450 (SOT PHE GLY+SCC GCL) | | Descriptor: | ENDOTHIAPEPSIN, N-[(1S)-2-{[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]amino}-1-(ethylsulfanyl)-2-oxoethyl]-Nalpha-(morpholin-4-ylsulfonyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Dealwis, C, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1EPR
 
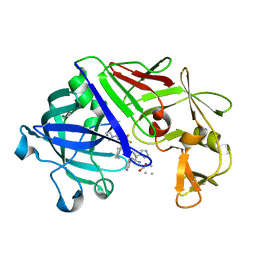 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-135,040 | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Badasso, M, Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EPS
 
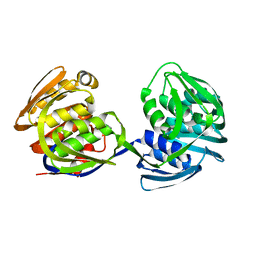 | | STRUCTURE AND TOPOLOGICAL SYMMETRY OF THE GLYPHOSPHATE 5-ENOL-PYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE: A DISTINCTIVE PROTEIN FOLD | | Descriptor: | 5-ENOL-PYRUVYL-3-PHOSPHATE SYNTHASE | | Authors: | Stallings, W.C, Abdel-Meguid, S.S, Lim, L.W, Shieh, H.-S, Dayringer, H.E, Leimgruber, N.K, Stegeman, R.A, Anderson, K.S, Sikorski, J.A, Padgette, S.R, Kishore, G.M. | | Deposit date: | 1991-04-05 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1EPT
 
 | | REFINED 1.8 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF PORCINE EPSILON-TRYPSIN | | Descriptor: | CALCIUM ION, PORCINE E-TRYPSIN | | Authors: | Huang, Q, Wang, Z, Li, Y, Liu, S, Tang, Y. | | Deposit date: | 1994-06-07 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A resolution crystal structure of the porcine epsilon-trypsin.
Biochim.Biophys.Acta, 1209, 1994
|
|
1EPU
 
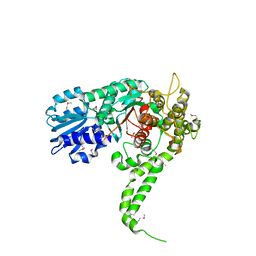 | | X-RAY crystal structure of neuronal SEC1 from squid | | Descriptor: | S-SEC1 | | Authors: | Bracher, A, Perrakis, A, Dresbach, T, Betz, H, Weissenhorn, W. | | Deposit date: | 2000-03-29 | | Release date: | 2000-08-09 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structure of neuronal Sec1 from squid sheds new light on the role of this protein in exocytosis.
Structure Fold.Des., 8, 2000
|
|
