6VC4
 
 | | Peanut lectin complexed with S-beta-D-Thiogalactopyranosyl beta-D-glucopyranoside derivative (STGD) | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6S)-3,4,5-trihydroxy-6-methoxytetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6E2B
 
 | | Ubiquitin in complex with Pt(2-phenilpyridine)(PPh3) | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin, ... | | Authors: | Zhemkov, V.A, Kim, M. | | Deposit date: | 2018-07-11 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reactions of Cyclometalated Platinum(II) [Pt(N∧C)(PR3)Cl] Complexes with Imidazole and Imidazole-Containing Biomolecules: Fine-Tuning of Reactivity and Photophysical Properties via Ligand Design.
Inorg Chem, 58, 2019
|
|
3H6T
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and cyclothiazide at 2.25 A resolution | | Descriptor: | ACETATE ION, CACODYLATE ION, CYCLOTHIAZIDE, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3UWZ
 
 | | Crystal structure of Staphylococcus aureus triosephosphate isomerase complexed with glycerol-2-phosphate | | Descriptor: | 2-HYDROXY-1-(HYDROXYMETHYL)ETHYL DIHYDROGEN PHOSPHATE, PHOSPHATE ION, Triosephosphate isomerase | | Authors: | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | Deposit date: | 2011-12-03 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
4H96
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-{3-[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine (UCP1018) | | Descriptor: | 5-{3-[3-(1,3-benzodioxol-5-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine, Dihydrofolate Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-09-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the active sites of dihydrofolate reductase from two species of Candida uncovers ligand-induced conformational changes shared among species.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6ASN
 
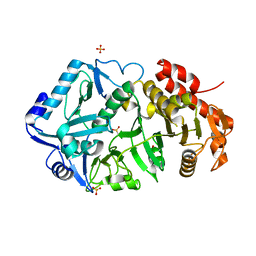 | |
6ASM
 
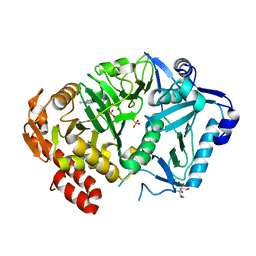 | | E. coli phosphoenolpyruvate carboxykinase G209S K212C mutant bound to thiosulfate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | Deposit date: | 2017-08-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
6AT4
 
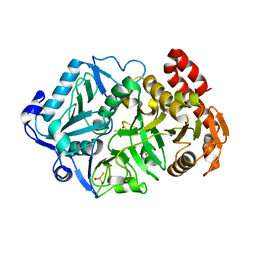 | |
4H97
 
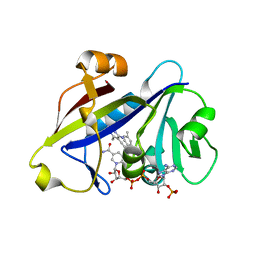 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5-(4-methylphenyl)phenyl]but-1-yn-1-yl}-6-methylpyrimidine-2,4-diamine (UCP111D4M) | | Descriptor: | 5-[(3R)-3-(5-methoxy-4'-methylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate Reductase, GLYCEROL, ... | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-09-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the active sites of dihydrofolate reductase from two species of Candida uncovers ligand-induced conformational changes shared among species.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5IZJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1411 | | Descriptor: | 4-(piperazin-1-yl)-7H-pyrrolo[2,3-d]pyrimidine, 47P-AZ1-DAR-DAR, 47P-AZ1-DAR-DAR-DAR, ... | | Authors: | Pflug, A, Enkvist, E, Uri, A, Engh, R.A. | | Deposit date: | 2016-03-25 | | Release date: | 2016-07-20 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bifunctional Ligands for Inhibition of Tight-Binding Protein-Protein Interactions.
Bioconjug.Chem., 27, 2016
|
|
6AT3
 
 | |
3H6W
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5217 at 1.50 A resolution | | Descriptor: | (3R)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
4H95
 
 | |
3UWV
 
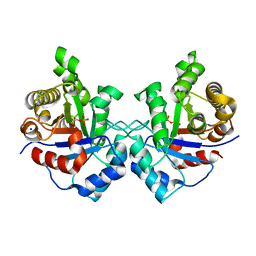 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with 2-phosphoglyceric acid | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, SODIUM ION, Triosephosphate isomerase | | Authors: | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | Deposit date: | 2011-12-03 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
6EU5
 
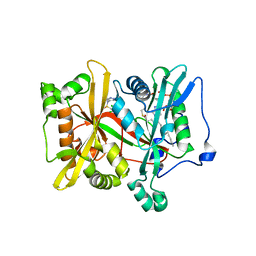 | | Leishmania major N-myristoyltransferase with bound myristoyl-CoA and inhibitor | | Descriptor: | 4-[3-[(8~{a}~{R})-3,4,6,7,8,8~{a}-hexahydro-1~{H}-pyrrolo[1,2-a]pyrazin-2-yl]propyl]-2,6-bis(chloranyl)-~{N}-methyl-~{N}-(1,3,5-trimethylpyrazol-4-yl)benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA | | Authors: | Brenk, R, Kehrein, J, Kersten, C. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.496083 Å) | | Cite: | How To Design Selective Ligands for Highly Conserved Binding Sites: A Case Study UsingN-Myristoyltransferases as a Model System.
J.Med.Chem., 2019
|
|
2P7Z
 
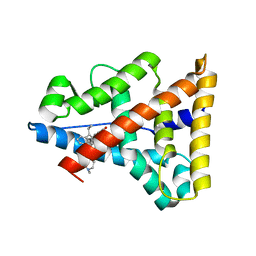 | | Estrogen Related Receptor Gamma in complex with 4-hydroxy-tamoxifen | | Descriptor: | 4-HYDROXYTAMOXIFEN, Estrogen-related receptor gamma | | Authors: | Abad, M.C. | | Deposit date: | 2007-03-21 | | Release date: | 2008-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determination of estrogen-related receptor gamma in the presence of phenol derivative compounds.
J.Steroid Biochem.Mol.Biol., 108, 2008
|
|
5IYS
 
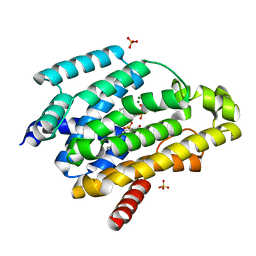 | | Crystal structure of a dehydrosqualene synthase in complex with ligand | | Descriptor: | MAGNESIUM ION, Phytoene synthase, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Liu, G.Z, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of a dehydrosqualene synthase in complex with ligand
to be published
|
|
6IPY
 
 | |
7LIR
 
 | | Structure of the invertebrate ALK GRD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALK tyrosine kinase receptor homolog scd-2, ... | | Authors: | Stayrook, S, Li, T, Klein, D.E. | | Deposit date: | 2021-01-27 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
7LRZ
 
 | | Structure of the Human ALK GRD | | Descriptor: | ALK tyrosine kinase receptor | | Authors: | Stayrook, S, Li, T, Klein, D.E. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
3GUI
 
 | |
3GUL
 
 | |
6M1I
 
 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
4WO0
 
 | | Crystal structure of transthyretin in complex with apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Folli, C, Berni, R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6IPZ
 
 | | Fyn SH3 domain R96W mutant, crystallized with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Tyrosine-protein kinase Fyn | | Authors: | Arold, S.T, Aljedani, S.S, Shahul Hameed, U.F. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | Synergy and allostery in ligand binding by HIV-1 Nef.
Biochem.J., 478, 2021
|
|
