4KA9
 
 | |
4DKY
 
 | | Crystal structure Analysis of N terminal region containing the dimerization domain and DNA binding domain of HU protein(Histone like protein-DNA binding) from Mycobacterium tuberculosis [H37Ra] | | Descriptor: | DNA-binding protein HU homolog, MANGANESE (II) ION | | Authors: | Bhowmick, T, Ramagopal, U.A, Ghosh, S, Nagaraja, V, Ramakumar, S. | | Deposit date: | 2012-02-05 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors
Nat Commun, 5, 2014
|
|
4K21
 
 | | Crystal structure of Canavalia boliviana lectin in complex with Xman | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Canavalia boliviana lectin, ... | | Authors: | Bezerra, G.A, Moura, T.R, Viertlmayr, R, Delatorre, P, Rocha, B.A.M, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2013-04-07 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of an Anti-Inflammatory Lectin from Canavalia boliviana Seeds in Complex with Dimannosides.
Plos One, 9, 2014
|
|
4K8C
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ADP | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ADP
To be Published
|
|
4DCE
 
 | | Crystal structure of human anaplastic lymphoma kinase in complex with a piperidine-carboxamide inhibitor | | Descriptor: | (3S)-N-(4-methylbenzyl)-1-{2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}piperidine-3-carboxamide, ALK tyrosine kinase receptor | | Authors: | Whittington, D.A, Epstein, L.F, Chen, H. | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rapid development of piperidine carboxamides as potent and selective anaplastic lymphoma kinase inhibitors.
J.Med.Chem., 55, 2012
|
|
4K2I
 
 | | Crystal structure of ntda from bacillus subtilis with bound cofactor pmp | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETATE ION, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2013-04-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4K93
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, N-(hydroxymethyl)benzamide, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 complexed with N-(HYDROXYMETHYL)BENZAMIDE
To be Published
|
|
4K30
 
 | | Structure of the N-acetyltransferase domain of human N-acetylglutamate synthase | | Descriptor: | N-ACETYL-L-GLUTAMATE, N-acetylglutamate synthase, mitochondrial | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of the N-acetyltransferase domain of human N-acetyl-L-glutamate synthase in complex with N-acetyl-L-glutamate provides insights into its catalytic and regulatory mechanisms.
Plos One, 8, 2013
|
|
4K3Z
 
 | |
4KBE
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with benzoguanamine | | Descriptor: | 6-phenyl-1,3,5-triazine-2,4-diamine, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with benzoguanamine
To be Published
|
|
4DCD
 
 | | 1.6A resolution structure of PolioVirus 3C Protease Containing a covalently bound dipeptidyl inhibitor | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Genome polyprotein, ... | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.-C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2012-01-17 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
4K5X
 
 | | Crystal structure of Staphylococcal nuclease variant V23M/L36F at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Caro, J.A, Flores, E, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-15 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pressure effects on proteins
To be Published
|
|
4KGE
 
 | | Crystal structure of near-infrared fluorescent protein with an extended stokes shift, pH 4.5 | | Descriptor: | CHLORIDE ION, TagRFP675, red fluorescent protein | | Authors: | Malashkevich, V.N, Piatkevich, K, Almo, S.C, Verkhusha, V, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 variant.
Sci Rep, 3, 2013
|
|
4K8T
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ethyl 3,4-diaminobenzoate | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with ethyl 3,4-diaminobenzoate
To be Published
|
|
4DMN
 
 | | HIV-1 Integrase Catalytical Core Domain | | Descriptor: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](methoxy)ethanoic acid, ARSENIC, HIV-1 Integrase, ... | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors.
J.Biol.Chem., 287, 2012
|
|
4DHS
 
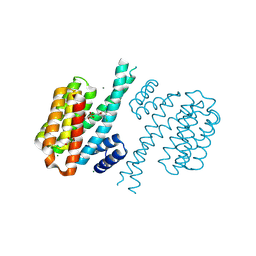 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3,5-dichlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 PROTEIN SIGMA, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DI1
 
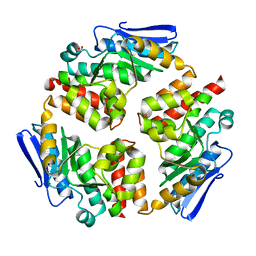 | |
4KBS
 
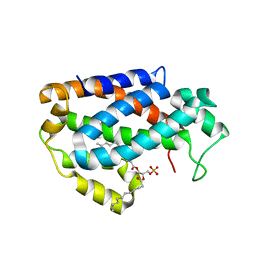 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 phosphatidic acid (12:0 PA) | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-ETHANEDIOL, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4DLP
 
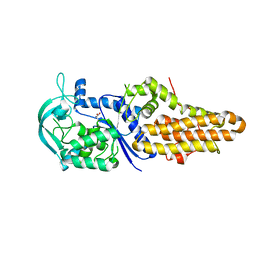 | |
4DN2
 
 | | CRYSTAL STRUCTURE OF putative Nitroreductase from Geobacter metallireducens GS-15 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF putative Nitroreductase from Geobacter metallireducens GS-15
To be Published
|
|
4DNG
 
 | | Crystal structure of putative aldehyde dehydrogenase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Uncharacterized aldehyde dehydrogenase AldY | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative aldehyde dehydrogenase from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
4K2H
 
 | | Crystal structure of C103A mutant of DJ-1 superfamily protein STM1931 from Salmonella typhimurium | | Descriptor: | Intracellular protease/amidase, ZINC ION | | Authors: | Shumilin, I.A, Niedzialkowska, E, Domagalski, M.J, Stam, J, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of C103A mutant of DJ-1 superfamily protein STM1931 from Salmonella typhimurium
TO BE PUBLISHED
|
|
4KGF
 
 | | Crystal structure of near-infrared fluorescent protein with an extended stokes shift, ph 8.0 | | Descriptor: | CHLORIDE ION, TagRFP675, red fluorescent protein | | Authors: | Malashkevich, V.N, Piatkevich, K, Almo, S.C, Verkhusha, V, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 variant.
Sci Rep, 3, 2013
|
|
4K4P
 
 | | TL-3 inhibited Trp6Ala HIV Protease | | Descriptor: | HIV-1 protease, NITRATE ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Tiefenbrunn, T, Stout, C.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic Fragment-Based Drug Discovery: Use of a Brominated Fragment Library Targeting HIV Protease.
Chem.Biol.Drug Des., 83, 2014
|
|
4KIO
 
 | | Kinase domain mutant of human Itk in complex with a covalently-binding inhibitor | | Descriptor: | 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]prop-2-en-1-one, 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2013-05-02 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of novel irreversible inhibitors of interleukin (IL)-2-inducible tyrosine kinase (Itk) by targeting cysteine 442 in the ATP pocket.
J.Biol.Chem., 288, 2013
|
|
