9EJE
 
 | |
7D79
 
 | | The structure of DcsB complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DltD domain-containing protein, ... | | Authors: | Tang, Y, Zhou, J.H, Wang, G.Q. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.10411429 Å) | | Cite: | A Polyketide Cyclase That Forms Medium-Ring Lactones.
J.Am.Chem.Soc., 143, 2021
|
|
5HJS
 
 | | Identification of LXRbeta selective agonists for the treatment of Alzheimer's Disease | | Descriptor: | 2-chloro-4-{1'-[(2R)-2-hydroxy-3-methyl-2-(trifluoromethyl)butanoyl]-4,4'-bipiperidin-1-yl}-N,N-dimethylbenzamide, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Parthasarathy, G, Klein, D. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Identification and in Vivo Evaluation of Liver X Receptor beta-Selective Agonists for the Potential Treatment of Alzheimer's Disease.
J.Med.Chem., 59, 2016
|
|
8EPM
 
 | | Human R-type voltage-gated calcium channel Cav2.3 CH2II-deleted mutant at 3.1 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-10-06 | | Release date: | 2022-12-14 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the R-type human Ca v 2.3 channel reveal conformational crosstalk of the intracellular segments.
Nat Commun, 13, 2022
|
|
5OT5
 
 | | The crystal structure of CK2alpha in complex with compound 24 | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
3R4K
 
 | |
8ERA
 
 | | RMC-5552 in complex with mTORC1 and FKBP12 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-5,9,27-trihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, 1-[6-{[(3M)-4-amino-3-(2-amino-1,3-benzoxazol-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-3,4-dihydroisoquinolin-2(1H)-yl]-3-hydroxypropan-1-one, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
7YKN
 
 | | Crystal structure of (6-4) photolyase from Vibrio cholerae | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cakilkaya, B, Kavakli, I.H, DeMirci, H. | | Deposit date: | 2022-07-23 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Vibrio cholerae (6-4) photolyase reveals interactions with cofactors and a DNA-binding region.
J.Biol.Chem., 299, 2023
|
|
3R8E
 
 | |
5IYP
 
 | |
5IYQ
 
 | |
7DB3
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP011-bound form | | Descriptor: | 4-bromanyl-2-[[2-[(E)-1-(3-methoxyphenyl)ethylideneamino]propan-2-ylamino]methyl]phenol, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
6FFF
 
 | | Human BRD2 C-terminal bromodomain with (S)-5-(1-acetyl-2-cyclopropyl-4-(2-(hydroxymethyl)benzyl)-1,2,3,4-tetrahydroquinoxalin-6-yl)pyrimidine-2-carboxamide | | Descriptor: | (S)-5-(1-acetyl-2-cyclopropyl-4-(2-(hydroxymethyl)benzyl)-1,2,3,4-tetrahydroquinoxalin-6-yl)pyrimidine-2-carboxamide, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Chung, C. | | Deposit date: | 2018-01-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of Tetrahydroquinoxalines as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the Second Bromodomain.
J.Med.Chem., 61, 2018
|
|
4MUU
 
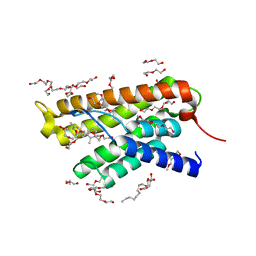 | | Structure of ThiT with pyrithiamine bound | | Descriptor: | 1-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-(2-HYDROXYETHYL)-2-METHYLPYRIDINIUM, 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Swier, L.J.Y.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2013-09-23 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies on the thiamin binding protein ThiT
To be Published
|
|
7XWZ
 
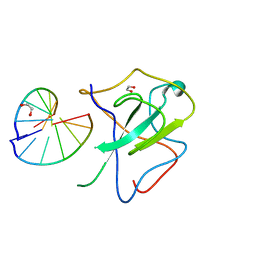 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
5V03
 
 | |
5WAD
 
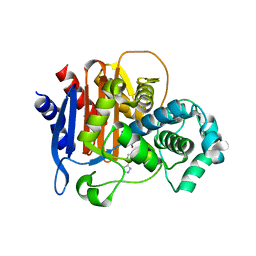 | | ADC-7 in complex with boronic acid transition state inhibitor CR161 | | Descriptor: | Beta-lactamase, PHOSPHATE ION, phosphonooxy-[[[6-(1~{H}-1,2,3,4-tetrazol-5-yl)pyridin-3-yl]sulfonylamino]methyl]borinic acid | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Based Analysis of Boronic Acids as Inhibitors of Acinetobacter-Derived Cephalosporinase-7, a Unique Class C beta-Lactamase.
ACS Infect Dis, 4, 2018
|
|
7HIT
 
 | | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with RA-0188454-02 (CHIKV_MacB-x2370) | | Descriptor: | 2-methyl-N-{[4-(2H-tetrazol-5-yl)-1,3-thiazol-2-yl]methyl}-1H-pyrrole-3-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Almahli, H, Balcomb, B.H, Capkin, E, Chandran, A.V, Chen, W, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Saleem, R.S.Z, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Todd, M.H, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center
To Be Published
|
|
8SD4
 
 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule fusion inhibitor compound 7 | | Descriptor: | (S~1~S)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-[(dimethylamino)methyl]azetidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Kadam, R.U, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7HIM
 
 | | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with RA-0188465-01 (CHIKV_MacB-x2188) | | Descriptor: | 2-(2-oxopyridin-1(2H)-yl)ethyl 3-methyl-1H-pyrrolo[2,3-b]pyridine-5-carboxylate, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Almahli, H, Balcomb, B.H, Capkin, E, Chandran, A.V, Chen, W, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Saleem, R.S.Z, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Todd, M.H, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center
To Be Published
|
|
5WNH
 
 | |
7AK1
 
 | | Human MALT1(329-729) in complex with a chromane urea containing inhibitor | | Descriptor: | 1-(3-chloranyl-4-methoxy-phenyl)-3-[7-[(3~{S})-3-(methoxymethyl)morpholin-4-yl]-2-methyl-pyrazolo[1,5-a]pyrimidin-6-yl]urea, MAGNESIUM ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Discovery of Potent, Highly Selective, and In Vivo Efficacious, Allosteric MALT1 Inhibitors by Iterative Scaffold Morphing.
J.Med.Chem., 63, 2020
|
|
7WFC
 
 | | X-ray structure of HKU1-PLP2(Cys109Ser) catalytic mutant in complex with free ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, 60S ribosomal protein L40, Papain-like protease, ... | | Authors: | Xiong, Y.X, Fu, Z.Y, Huang, H. | | Deposit date: | 2021-12-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The substrate selectivity of papain-like proteases from human-infecting coronaviruses correlates with innate immune suppression.
Sci.Signal., 16, 2023
|
|
2W4F
 
 | | CRYSTAL STRUCTURE OF THE FIRST PDZ DOMAIN OF HUMAN SCRIB1 | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN LAP4 | | Authors: | Hozjan, V, Pilka, E.S, Roos, A.K, W Yue, W, Phillips, C, Bray, J, Cooper, C, Salah, E, Elkins, J.M, Muniz, J.R.C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Bountra, C, Doyle, D.A, Oppermann, U. | | Deposit date: | 2008-11-25 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the First Pdz Domain of Human Scrib1
To be Published
|
|
5CGB
 
 | | Crystal structure of FimH in complex with heptyl alpha-D-septanoside | | Descriptor: | (6R)-1,6-anhydro-2-O-heptyl-6-(hydroxymethyl)-D-galactitol, Protein FimH, SULFATE ION | | Authors: | Jakob, R.P, Preston, R.P, Zihlmann, P, Fiege, B, Sager, C.P, Vannam, R, Rabbani, S, Zalewski, A, Maier, T, Ernst, B, Peczuh, M. | | Deposit date: | 2015-07-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The price of flexibility - a case study on septanoses as pyranose mimetics.
Chem Sci, 9, 2018
|
|
