7XIF
 
 | | Crystal structure of the aminopropyltransferase, SpeE from hyperthermophilic crenarchaeon, Pyrobaculum calidifontis in complex with 5'-methylthioadenosine (MTA) alone or together with spermidine or thermospermine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE, Polyamine aminopropyltransferase, ... | | Authors: | Mizohata, E, Yasuda, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Substrate Specificity of an Aminopropyltransferase and the Biosynthesis Pathway of Polyamines in the Hyperthermophilic Crenarchaeon Pyrobaculum calidifontis.
Catalysts, 12, 2022
|
|
6A3A
 
 | | MVM NES mutant Nm2 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, Exportin-1, GLYCEROL, ... | | Authors: | Sun, Q, Li, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cancer Therapy with Nanoparticle-Medicated Intracellular Expression of Peptide CRM1-Inhibitor.
Int J Nanomedicine, 16, 2021
|
|
6DR0
 
 | | Class 5 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5BYT
 
 | |
6JX1
 
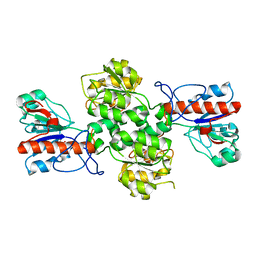 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101 | | Descriptor: | Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
3GWC
 
 | |
7BWC
 
 | |
8HQF
 
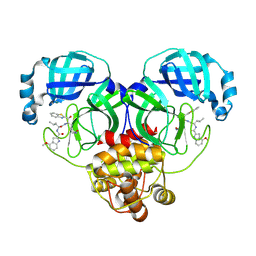 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Non-structural protein 7 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2024-01-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
5SUF
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03H03 from the F2X-Universal Library | | Descriptor: | 1-benzyl-1H-pyrazole-4-carboxylic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
1MXJ
 
 | | NMR solution structure of benz[a]anthracene-dG in ras codon 12,2; GGCAGXTGGTG | | Descriptor: | 1S,2R,3S,4R-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*AP*CP*CP*AP*CP*CP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*G)-3' | | Authors: | Kim, H.-Y.H, Wilkinson, A.S, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minor Groove Orientation for the (1S,2R,3S,4R)-N2-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxy-benz[a]anthracenyl)]-2'-deoxyguanosyl Adduct in the N-ras Codon 12 sequence
Biochemistry, 42, 2003
|
|
5STC
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P02F09 from the F2X-Universal Library | | Descriptor: | 1-(2-chloroethyl)-1H-imidazole, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5C84
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 20 | | Descriptor: | (2R,5R)-4-[2-(6-chloro-3,3-dimethyl-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-5-(methoxymethyl)-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
7FHP
 
 | |
3H10
 
 | | Aurora A inhibitor complex | | Descriptor: | 9-chloro-7-(2,6-difluorophenyl)-N-{4-[(4-methylpiperazin-1-yl)carbonyl]phenyl}-5H-pyrimido[5,4-d][2]benzazepin-2-amine, Serine/threonine-protein kinase 6 | | Authors: | Wiesmann, C, Ultsch, M.H, Cochran, A.G. | | Deposit date: | 2009-04-10 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B.
J.Med.Chem., 52, 2009
|
|
5C01
 
 | | Crystal Structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NON-RECEPTOR TYROSINE-PROTEIN KINASE TYK2, ... | | Authors: | Min, X, Wang, Z, Walker, N. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of the JH2 Pseudokinase Domain of JAK Family Tyrosine Kinase 2 (TYK2).
J.Biol.Chem., 290, 2015
|
|
7I9X
 
 | | Group deposition of ZIKV NS2B-NS3 protease in complex with inhibitors from ASAP Discovery Consortium -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with ASAP-0029394-001 | | Descriptor: | DIMETHYL SULFOXIDE, N-(2,3-dihydro-1H-isoindol-5-yl)-6-fluoro-1H-1,2,3-benzotriazole-4-carboxamide, Serine protease NS3, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Lee, A, Kenton, N, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2025-04-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Group deposition of ZIKV NS2B-NS3 protease in complex with inhibitors from ASAP Discovery Consortium
To Be Published
|
|
3DYH
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-721 | | Descriptor: | 3-butoxy-1-(2,2-diphosphonoethyl)pyridinium, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2008-07-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
4YQS
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | (5-amino-1H-1,2,4-triazol-1-yl)(4-methoxyphenyl)methanone, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
4YRE
 
 | |
6K10
 
 | | Non substrate bound state of Staphylococcus Aureus AldH | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase | | Authors: | Zhang, Z, Tao, X. | | Deposit date: | 2019-05-08 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78962183 Å) | | Cite: | Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase
CCS Chemistry, 2, 2020
|
|
8VVT
 
 | | Mammalian ribosomes bound to Anisomycin in the rotated conformation | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Loerch, S, Petrossian, E, Smith, P.R, Campbell, Z.T. | | Deposit date: | 2024-01-31 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid purification reveals structural insights into transient ribosome states
To Be Published
|
|
4RFM
 
 | | ITK kinase domain in complex with compound 1 N-{1-[(1,1-dioxo-1-thian-2-yl)(phenyl)methyl]-1H- pyrazol-4-yl}-5,5-difluoro-5a-methyl-1H,4H,4aH,5H,5aH,6H-cyclopropa[f]indazole-3-carboxamide | | Descriptor: | (4aS,5aR)-N-{1-[(R)-[(2R)-1,1-dioxidotetrahydro-2H-thiopyran-2-yl](phenyl)methyl]-1H-pyrazol-4-yl}-5,5-difluoro-5a-methyl-1,4,4a,5,5a,6-hexahydrocyclopropa[f]indazole-3-carboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | McEwan, P.A, Barker, J.J, Eigenbrot, C. | | Deposit date: | 2014-09-26 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetrahydroindazoles as Interleukin-2 Inducible T-Cell Kinase Inhibitors. Part II. Second-Generation Analogues with Enhanced Potency, Selectivity, and Pharmacodynamic Modulation in Vivo.
J.Med.Chem., 58, 2015
|
|
8ILV
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 18 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[3-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6DYV
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-((pent-4-yn-1-ylthio)methyl)pyrrolidin-3-ol | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(pent-4-yn-1-yl)sulfanyl]methyl}pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2018-07-02 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
3DFT
 
 | |
