6ULG
 
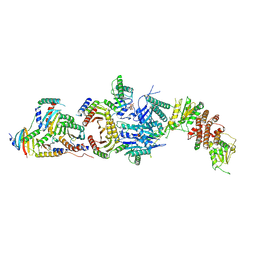 | | Cryo-EM structure of the FLCN-FNIP2-Rag-Ragulator complex | | Descriptor: | Folliculin, Folliculin-interacting protein 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shen, K, Rogala, K.B, Yu, Z.H, Sabatini, D.M. | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM Structure of the Human FLCN-FNIP2-Rag-Ragulator Complex.
Cell, 179, 2019
|
|
6P5W
 
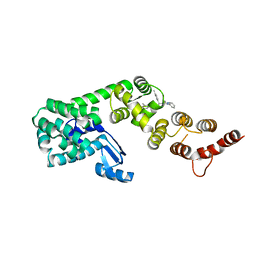 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6PGN
 
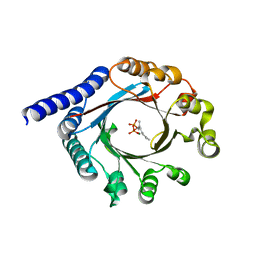 | | PagF single mutant with GPP | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, PagF | | Authors: | Nair, S.K, Hao, Y, Estrada, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Single Amino Acid Switch Alters the Isoprene Donor Specificity in
Ribosomally Synthesized and Post-Translationally Modified Peptide
Prenyltransferases
J. Am. Chem. Soc., 140, 2018
|
|
6P5V
 
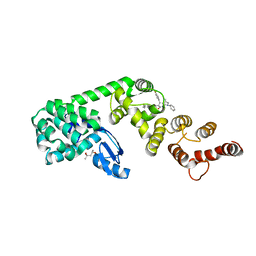 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6P6I
 
 | | Structure of YbtPQ importer | | Descriptor: | ABC transporter protein, inner membrane ABC-transporter | | Authors: | Wang, Z, Hu, W, Zheng, H. | | Deposit date: | 2019-06-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter.
Sci Adv, 6, 2020
|
|
6PGM
 
 | | PirF geranyltransferase | | Descriptor: | PirF Geranyltransferase | | Authors: | Nair, S.K, Hao, Y, Estrada, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Single Amino Acid Switch Alters the Isoprene Donor Specificity in Ribosomally Synthesized and Post-Translationally Modified Peptide Prenyltransferases.
J.Am.Chem.Soc., 140, 2018
|
|
5NTW
 
 | |
5NU1
 
 | |
6VLZ
 
 | | Structure of the human mitochondrial ribosome-EF-G1 complex (ClassI) | | Descriptor: | 12s rRNA, 16s rRNA, 28S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Koripella, R.K, Agrawal, R.K. | | Deposit date: | 2020-01-27 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of the human mitochondrial ribosome bound to EF-G1 reveal distinct features of mitochondrial translation elongation.
Nat Commun, 11, 2020
|
|
2N9E
 
 | |
6QDY
 
 | | The crystal structure of Sporosarcina pasteurii urease in complex with its substrate urea | | Descriptor: | 1,2-ETHANEDIOL, FLUORIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Benini, S, Ciurli, S. | | Deposit date: | 2019-01-03 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | The Structure of the Elusive Urease-Urea Complex Unveils the Mechanism of a Paradigmatic Nickel-Dependent Enzyme.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8V83
 
 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
7LVS
 
 | | The CBP TAZ1 Domain in Complex with a CITED2-HIF-1-Alpha Fusion Peptide | | Descriptor: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone lysine acetyltransferase CREBBP, ZINC ION | | Authors: | Appling, F.D, Berlow, R.B, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The molecular basis of allostery in a facilitated dissociation process.
Structure, 29, 2021
|
|
5NTN
 
 | | Structural states of RORgt: X-ray elucidation of molecular mechanisms and binding interactions for natural and synthetic compounds | | Descriptor: | (5R,10S,13R,14R,17R)-17-((R,E)-7-hydroxy-6-methylhept-5-en-2-yl)-4,4,10,13,14-pentamethyl-1,2,5,6,10,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthrene-3,7(4H)-dione, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Kallen, J. | | Deposit date: | 2017-04-28 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural States of ROR gamma t: X-ray Elucidation of Molecular Mechanisms and Binding Interactions for Natural and Synthetic Compounds.
ChemMedChem, 12, 2017
|
|
5KEI
 
 | |
5JA2
 
 | | EntF, a Terminal Nonribosomal Peptide Synthetase Module Bound to the non-Native MbtH-Like Protein PA2412 | | Descriptor: | 5'-({[(2R,3S)-3-amino-4-hydroxy-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}butyl]sulfonyl}amino)-5'-deoxyadenosine, Enterobactin synthase component F, MbtH-Like Protein PA2412, ... | | Authors: | Miller, B.R, Gulick, A.M. | | Deposit date: | 2016-04-11 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a Nonribosomal Peptide Synthetase Module Bound to MbtH-like Proteins Support a Highly Dynamic Domain Architecture.
J.Biol.Chem., 291, 2016
|
|
8OIR
 
 | | 55S human mitochondrial ribosome with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
6FG7
 
 | | Crystal structure of the BIR2 ectodomain from Arabidopsis thaliana. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Inactive LRR receptor-like serine/threonine-protein kinase BIR2, ... | | Authors: | Hothorn, M, Hohmann, U. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling.
Nat Plants, 4, 2018
|
|
6WJ3
 
 | |
5J1S
 
 | | TorsinA-LULL1 complex, H. sapiens, bound to VHH-BS2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Demircioglu, F.E, Schwartz, T.U. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia.
Elife, 5, 2016
|
|
6WJ2
 
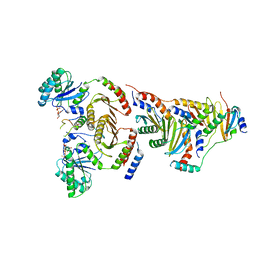 | | CryoEM structure of the SLC38A9-RagA-RagC-Ragulator complex in the pre-GAP state | | Descriptor: | 9-{5-O-[(S)-hydroxy{[(R)-hydroxy(thiophosphonooxy)phosphoryl]oxy}phosphoryl]-alpha-L-arabinofuranosyl}-3,9-dihydro-1H-purine-2,6-dione, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fromm, S.A, Hurley, J.H. | | Deposit date: | 2020-04-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism for amino acid-dependent Rag GTPase nucleotide state switching by SLC38A9.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6FQP
 
 | |
6FQQ
 
 | |
