7HK3
 
 | |
4PQ0
 
 | | Crystal structure of POB3 middle domain at 1.65A | | Descriptor: | FACT complex subunit POB3, MALONIC ACID | | Authors: | Liu, H. | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of POB3 middle domain at 1.65A
TO BE PUBLISHED
|
|
5EUU
 
 | | Rat prestin STAS domain in complex with chloride | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Prestin,Prestin, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
4JJH
 
 | | Crystal structure of the D1 domain from human Nectin-4 extracellular fragment [PSI-NYSGRC-005624] | | Descriptor: | 1,2-ETHANEDIOL, Poliovirus receptor-related protein 4 | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-07 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D1 domain of human Nectin-4 extracellular fragment [NYSGRC-005624]
to be published
|
|
5EUS
 
 | | Rat prestin STAS domain in complex with bromide | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Prestin,Rat prestin STAS domain, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5EUW
 
 | | Rat prestin STAS domain in complex with nitrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NITRATE ION, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5EUX
 
 | | Rat prestin STAS domain in complex with thiocyanate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Prestin,Prestin, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.038 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
1BTX
 
 | |
6MNU
 
 | | Crystal structure of Yersinia pestis UDP-glucose pyrophosphorylase | | Descriptor: | 1,2-ETHANEDIOL, UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Gibbs, M.E, Lountos, G.T, Gumpena, R, Waugh, D.S. | | Deposit date: | 2018-10-03 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Crystal structure of UDP-glucose pyrophosphorylase from Yersinia pestis, a potential therapeutic target against plague.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5EUZ
 
 | | Rat prestin STAS domain in complex with iodide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IODIDE ION, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5F1Q
 
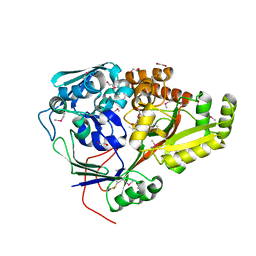 | | Crystal Structure of Periplasmic Dipeptide Transport Protein from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-30 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Crystal Structure of Periplasmic Dipeptide Transport Protein from Yersinia pestis
To Be Published
|
|
4H79
 
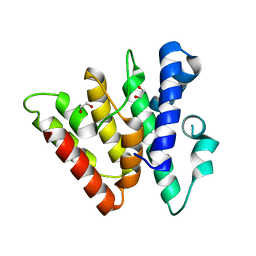 | | Crystal structure of CasB from Thermobifida fusca | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated protein, Cse2 family | | Authors: | Ke, A, Nam, K.H. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures.
Febs Lett., 586, 2012
|
|
1XKW
 
 | | Pyochelin outer membrane receptor FptA from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Fe(III)-pyochelin receptor, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Cobessi, D, Celia, H, Pattus, F. | | Deposit date: | 2004-09-30 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa
J.Mol.Biol., 352, 2005
|
|
2XCE
 
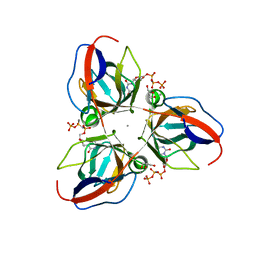 | | Structure of YncF in complex with dUpNHpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Garcia-Nafria, J, Burchell, L, Takezawa, M, Rzechorzek, N, Fogg, M, Wilson, K.S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Genomic Bacillus Subtilis Dutpase: Novel Features in the Phe-Lid.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1C2A
 
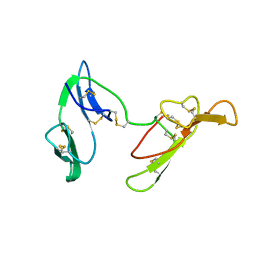 | | CRYSTAL STRUCTURE OF BARLEY BBI | | Descriptor: | BOWMAN-BIRK TRYPSIN INHIBITOR | | Authors: | Song, H.K, Kim, Y.S, Yang, J.K, Moon, J, Lee, J.Y, Suh, S.W. | | Deposit date: | 1999-07-23 | | Release date: | 1999-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 A resolution.
J.Mol.Biol., 293, 1999
|
|
4LWH
 
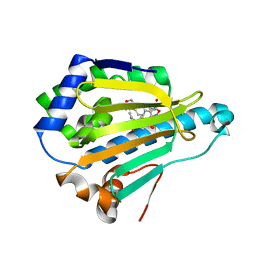 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LZB
 
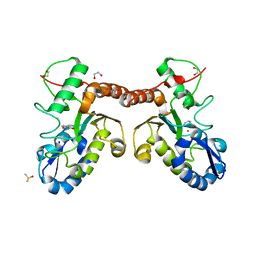 | | Uracil binding pocket in Vaccinia virus uracil DNA glycosylase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the uracil complex of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5VNB
 
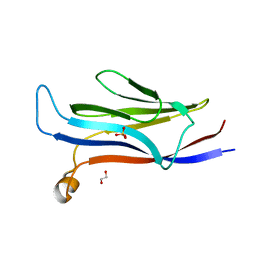 | | YEATS in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, H3K23acK27ac peptide, SULFATE ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2017-04-29 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | GAS41 Recognizes Diacetylated Histone H3 through a Bivalent Binding Mode.
ACS Chem. Biol., 13, 2018
|
|
1C44
 
 | |
1XCS
 
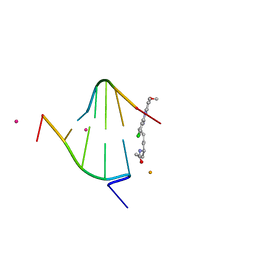 | | structure of oligonucleotide/drug complex | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-[(5-(ACETYLAMINO)-6-{[(1S,4R)-8-AMINO-4-[((2R)-6-AMINO-2-{2-[(1S)-5-AMINO-1-FORMYLPENTYL]HYDRAZINO}HEXANOYL)AMINO]-1-(4-AMINOBUTYL)-2,3-DIOXOOCTYL]AMINO}-6-OXOHEXYL)AMINO]-6-CHLORO-2-METHOXYACRIDINIUM, BARIUM ION, ... | | Authors: | Valls, N, Steiner, R.A, Wright, G, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-09-03 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Variable role of ions in two drug intercalation complexes of DNA
J.Biol.Inorg.Chem., 10, 2005
|
|
4JTB
 
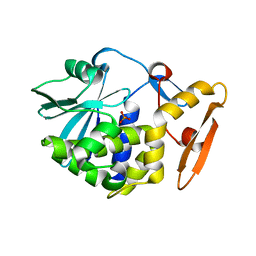 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with phosphate ion at 1.71 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, rRNA N-glycosidase | | Authors: | Pandey, S, Tyagi, T.K, Singh, A, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-23 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with phosphate ion at 1.71 Angstrom resolution
To be published
|
|
4MXN
 
 | |
4BCJ
 
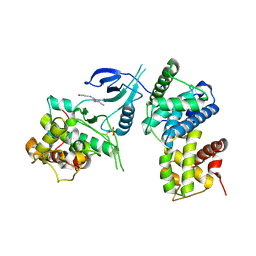 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[(3-hydroxyphenyl)amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1 | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
1XSN
 
 | | Crystal Structure of human DNA polymerase lambda in complex with a one nucleotide DNA gap and ddTTP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*(2DT))-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2YRA
 
 | |
