1EE9
 
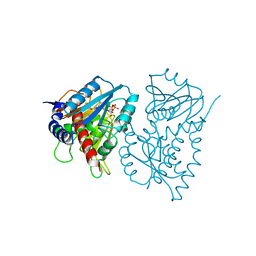 | | CRYSTAL STRUCTURE OF THE NAD-DEPENDENT 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH NAD | | Descriptor: | 5,10-METHYLENETETRAHYDROFOLATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Monzingo, A.F, Breksa, A, Ernst, S, Appling, D.R, Robertus, J.D. | | Deposit date: | 2000-01-31 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray structure of the NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Saccharomyces cerevisiae.
Protein Sci., 9, 2000
|
|
1EEA
 
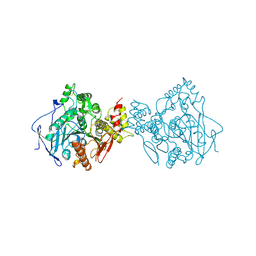 | | Acetylcholinesterase | | Descriptor: | PROTEIN (ACETYLCHOLINESTERASE) | | Authors: | Raves, M.L, Giles, K, Schrag, J.D, Schmid, M.F, Phillips Jr, G.N, Wah, C, Howard, A.J, Silman, I, Sussman, J.L. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary Structure of Tetrameric Acetylcholinesterase
Structure and Function of Cholinesterases and Related Proteins, 1998
|
|
1EED
 
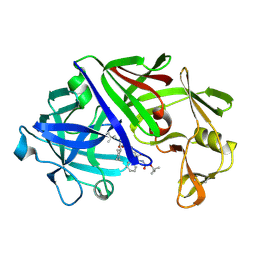 | | X-ray crystallographic analysis of inhibition of endothiapepsin by cyclohexyl renin inhibitors | | Descriptor: | (2S)-2-[[(3S,4S)-5-cyclohexyl-4-[[(4S,5S)-5-[(2-methylpropan-2-yl)oxycarbonylamino]-4-oxidanyl-6-phenyl-hexanoyl]amino] -3-oxidanyl-pentanoyl]amino]-4-methyl-pentanoic acid, ENDOTHIAPEPSIN | | Authors: | Blundell, T.L, Frazao, C, Cooper, J.B. | | Deposit date: | 1992-06-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic analysis of inhibition of endothiapepsin by cyclohexyl renin inhibitors.
Biochemistry, 31, 1992
|
|
1EEF
 
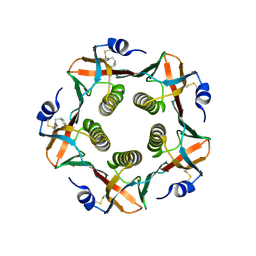 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER COMPLEXED WITH BOUND LIGAND PEPG | | Descriptor: | 2-PHENETHYL-2,3-DIHYDRO-PHTHALAZINE-1,4-DIONE, PROTEIN (HEAT-LABILE ENTEROTOXIN B CHAIN), alpha-D-galactopyranose | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the GM1 receptor-binding site of heat-labile enterotoxin and cholera toxin by phenyl-ring-containing galactose derivatives.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EEG
 
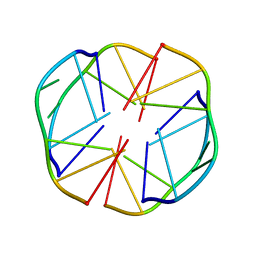 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
1EEH
 
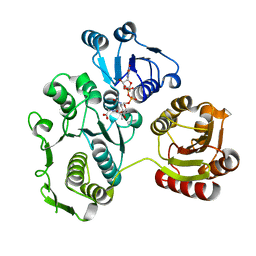 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
1EEI
 
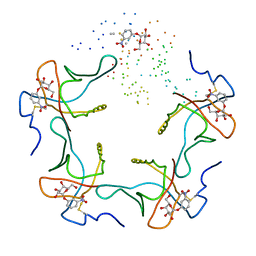 | |
1EEJ
 
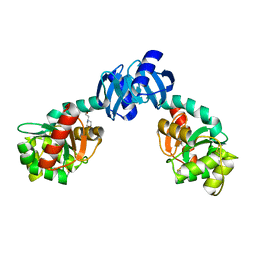 | | CRYSTAL STRUCTURE OF THE PROTEIN DISULFIDE BOND ISOMERASE, DSBC, FROM ESCHERICHIA COLI | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, THIOL:DISULFIDE INTERCHANGE PROTEIN | | Authors: | McCarthy, A.A, Haebel, P.W, Torronen, A, Rybin, V, Baker, E.N, Metcalf, P. | | Deposit date: | 2000-01-31 | | Release date: | 2000-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli.
Nat.Struct.Biol., 7, 2000
|
|
1EEK
 
 | | SOLUTION STRUCTURE OF A NONPOLAR, NON HYDROGEN BONDED BASE PAIR SURROGATE IN DNA. | | Descriptor: | 5'-D(*CP*GP*CP*AP*TP*(DFT)P*GP*TP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*AP*CP*(MBZ)P*AP*TP*GP*CP*G)-3' | | Authors: | Kool, E.T, Krugh, T.R, Guckian, K.M. | | Deposit date: | 2000-02-01 | | Release date: | 2000-02-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Nonpolar, Non-Hydrogen-Bonded Base Pair Surrogate in DNA
J.Am.Chem.Soc., 122, 2000
|
|
1EEL
 
 | |
1EEM
 
 | | GLUTATHIONE TRANSFERASE FROM HOMO SAPIENS | | Descriptor: | GLUTATHIONE, GLUTATHIONE-S-TRANSFERASE, SULFATE ION | | Authors: | Board, P, Coggan, M, Chelvanayagam, G, Easteal, S, Jermiin, L.S, Schulte, G.K, Danley, D.E, Hoth, L.R, Griffor, M.C, Kamath, A.V, Rosner, M.H, Chrunyk, B.A, Perregaux, D.E, Gabel, C.A, Geoghegan, K.F, Pandit, J. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and crystal structure of the Omega class glutathione transferases.
J.Biol.Chem., 275, 2000
|
|
1EEN
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-D-A-D-BPA-PTYR-L-I-P-Q-Q-G | | Descriptor: | ACETIC ACID, ALA-ASP-PBF-PTR-LEU-ILE-PRO, MAGNESIUM ION, ... | | Authors: | Puius, Y.A, Zhao, Y, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
1EEO
 
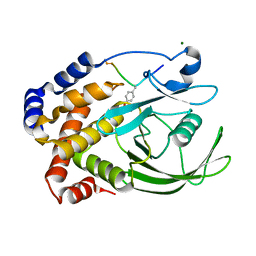 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 | | Descriptor: | ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 PEPTIDE, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Sarmiento, M, Puius, Y.A, Vetter, S.W, Lawrence, D.S, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
1EEP
 
 | | 2.4 A RESOLUTION CRYSTAL STRUCTURE OF BORRELIA BURGDORFERI INOSINE 5'-MONPHOSPHATE DEHYDROGENASE IN COMPLEX WITH A SULFATE ION | | Descriptor: | INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | McMillan, F.M, Cahoon, M, White, A, Hedstrom, L, Petsko, G.A, Ringe, D. | | Deposit date: | 2000-02-01 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 A resolution of Borrelia burgdorferi inosine 5'-monophosphate dehydrogenase: evidence of a substrate-induced hinged-lid motion by loop 6.
Biochemistry, 39, 2000
|
|
1EEQ
 
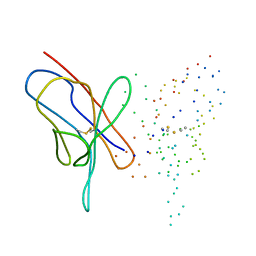 | | M4L/Y(27D)D/T94H Mutant of LEN | | Descriptor: | KAPPA-4 IMMUNOGLOBULIN (LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Raffen, R, Dieckman, L, Boogaard, C, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Increasing protein stability by polar surface residues: domain-wide consequences of interactions within a loop.
Biophys.J., 82, 2002
|
|
1EER
 
 | |
1EES
 
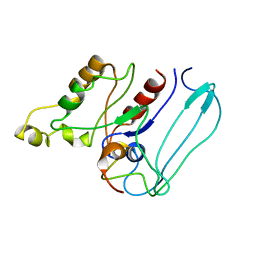 | | SOLUTION STRUCTURE OF CDC42HS COMPLEXED WITH A PEPTIDE DERIVED FROM P-21 ACTIVATED KINASE, NMR, 20 STRUCTURES | | Descriptor: | GTP-BINDING PROTEIN, P21-ACTIVATED KINASE | | Authors: | Gizachew, D, Guo, W, Chohan, K.C, Sutcliffe, M.J, Oswald, R.E. | | Deposit date: | 2000-02-02 | | Release date: | 2000-03-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of Cdc42Hs with a peptide derived from P-21 activated kinase.
Biochemistry, 39, 2000
|
|
1EET
 
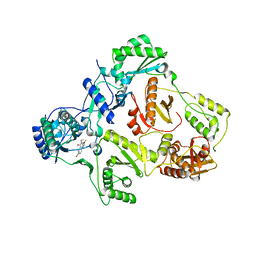 | | HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH THE INHIBITOR MSC204 | | Descriptor: | 1-(5-BROMO-PYRIDIN-2-YL)-3-[2-(6-FLUORO-2-HYDROXY-3-PROPIONYL-PHENYL)-CYCLOPROPYL]-UREA, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Hogberg, M, Sahlberg, C, Engelhardt, P, Noreen, R, Kangasmetsa, J, Johansson, N.G, Oberg, B, Vrang, L, Zhang, H, Sahlberg, B.L, Unge, T, Lovgren, S, Fridborg, K, Backbro, K. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues.
J.Med.Chem., 42, 1999
|
|
1EEU
 
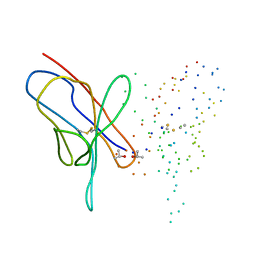 | | M4L/Y(27D)D/Q89D/T94H mutant of LEN | | Descriptor: | ISOPROPYL ALCOHOL, KAPPA-4 IMMUNOGLOBULIN (LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Cai, X, Gu, M, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Factors contributing to decreased protein stability when aspartic acid residues are in beta-sheet regions.
Protein Sci., 11, 2002
|
|
1EEX
 
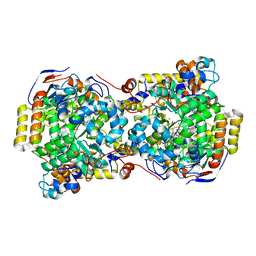 | | CRYSTAL STRUCTURE OF THE DIOL DEHYDRATASE-ADENINYLPENTYLCOBALAMIN COMPLEX FROM KLEBSIELLA OXYTOCA | | Descriptor: | CO-(ADENIN-9-YL-PENTYL)-COBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | Authors: | Shibata, N, Masuda, J, Toraya, T, Morimoto, Y, Yasuoka, N. | | Deposit date: | 2000-02-04 | | Release date: | 2001-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
1EEY
 
 | |
1EEZ
 
 | |
1EF0
 
 | |
1EF1
 
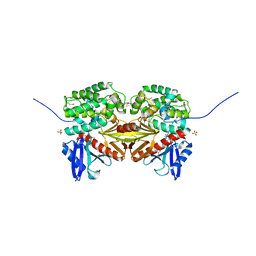 | | CRYSTAL STRUCTURE OF THE MOESIN FERM DOMAIN/TAIL DOMAIN COMPLEX | | Descriptor: | MOESIN, SULFATE ION | | Authors: | Pearson, M.A, Reczek, D, Bretscher, A, Karplus, P.A. | | Deposit date: | 2000-02-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain.
Cell(Cambridge,Mass.), 101, 2000
|
|
1EF2
 
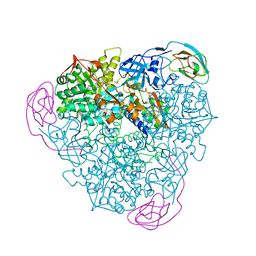 | | CRYSTAL STRUCTURE OF MANGANESE-SUBSTITUTED KLEBSIELLA AEROGENES UREASE | | Descriptor: | MANGANESE (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Yamaguchi, K, Cosper, N.J, Stalhandske, C, Scott, R.A, Pearson, M.A, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-02-05 | | Release date: | 2000-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of metal-substituted Klebsiella aerogenes urease.
J.Biol.Inorg.Chem., 4, 1999
|
|
